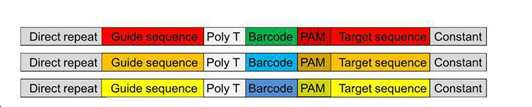
METHOD OF IN VIVO EVALUATION OF RNA-GUIDED NUCLEASE ACTIVITY IN HIGH-THROUGHPUT MANNER
The present invention refers to (Clustered regularly interspaced short palindromic repeats and CRISPR non-associated protein) derived from RNA - guide new the [ley which will grow oh number represented by the number 2 type CRISPR-a Cas (RNA-a guided nuclease) immune system such that only one is used means [...] number to other. In particular, cellular proteins using single guide RNA (sgRNA) on inserting a dielectric and organ Cas9 technique associated with immunohistochemical studies aluminum (Cell, 2014, 157:1262 - 1278). In particular, a single guide RNA (single guide RNA, sgRNA) to predict the activity of CRISPR provided Cas9 system studies and progressing (ACS Synth Biol. , 2017, Feb 10; Sci Rep, 2016, 6:30870, Nat Biotechnol, 34, 184 - 191), and CRISPR provided Cas9 gene encoding patient lung cancer cells for reparing over by PD-a 1 number is a property, for the treatment of diseases using CRISPR provided Cas9 be progressing disclosed (Nature, 2016, 539:479) study in China. Recently, Cpf1 protein ( Dielectric correction number but it is highly important new the [ley which will grow oh guide RNA - applying thereof in active and accuracy, target and non-target activity uses a short code number RNA - guide new the [ley which will grow oh efforts were disclosed. , the victims of the present invention The aim of the one of the present invention new the [ley which will grow oh number (a) (RNA-a guided nuclease) have been introduced RNA - guide, guide RNA (guide RNA) encoding a desired target bio-sequence listing including said guide RNA oligonucleotides including bio-sequence listing and is obtained from a library cells separated using performing DNA sequence analysis; and (B) target sequence pair (pair) of respective guide from the data obtained through said sequencing RNA - is and it is burnt (indel frequency) including a frequency of detecting, evaluating the activity of a new the [ley which will grow oh RNA - guide number (RNA-a guided nuclease) number [...] method are disclosed. It is another object of the present invention guide RNA encoding a desired target RNA oligonucleotides including cells including bio-sequence listing bio-sequence listing and said guide is at least one number 2 including [...] library cells are disclosed. It is another object of the present invention guide RNA encoding a desired target RNA oligonucleotides including bio-sequence listing and said guide is separated bio-sequence listing including vector, and vector library [...] number are disclosed. It is another object of the present invention guide RNA encoding a desired target RNA oligonucleotide bio-sequence listing including bio-sequence listing and said guide is separated, and oligonucleotide library [...] number are disclosed. It is another object of the present invention (a) involves setting a target bio-sequence listing target RNA - guide new the [ley which will grow oh [thing it does, character number; (b) setting said target bio-sequence listing complementary chain with a base forming a gear pair, guide RNA nucleotide sequence encoding design step; (c) said guide RNA oligonucleotide including a designing step bio-sequence listing the purpose and target; said step (a) to (c) and (d) repeating step 1 or more times including including, a method for constructing a number [...] oligonucleotide library are disclosed. It is another object of the present invention in PAM (proto-a spacer provided adjacent Motif) sequence CTTA TTTV or adjacent to, target bio-sequence listing complementary chain (complementary strand) with a base sequence including the gear forming a gear pair, a separate guide RNA [...] number are disclosed. It is another object of the present invention separated said guide RNA, or nucleic acids encoding the same including, for correcting dielectric composition [...] number are disclosed. It is another object of the present invention separated said guide RNA, or nucleic acid encoding the same; and Cpf1 protein or nucleic acid encoding the same including, in a mammalian cell system for calibrating a dielectric [...] number are disclosed. It is another object of the present invention said guide RNA or nucleic acid encoding the same; and Cpf1 protein or nucleic acid encoding the same isolated mammalian cell including sequentially or simultaneously introducing, in a mammalian cell a number [...] Cpf1 dielectric correction in an optical method are disclosed. (Programmable nuclease) which is widely used and locked dielectric calibration cells scissors gene, said gene scissors calibration technique is using same life science, inner bio-technology and medicinal applications suitable metals and Sn-SB can be used. In particular, immune system derived RNA - new the [ley which will grow oh number number 2 type CRISPR/Cas cookies (clustered regularly interspaced repeat/CRISPR non-associated) (RNA-a guided nuclease) such as prokaryotic obtained in its usefulness to Cpf1 Cas9 and guide the spotlight disclosed. Only, said RNA - guide new the [ley which will grow oh for utilization of an RNA target sequence number order number new the [ley which will grow oh which is designed such that a guide for which it is highly important, this guide is active (on a non-target activity) target and non-target RNA has the nucleotide sequence according may change's oldest active (off-a target activity). , the victims of the activity of the present invention new the [ley which will grow oh number RNA - guide And specifically described as follows. On the other hand, in the present invention disclosure form embodiment described and each form can be applied to each of the other described and embodiment. I.e., various elements of the present invention combination in the present invention disclosure all into the same category. In addition, the technique of the present invention is described specifically by number one category is that it becomes, ball cannot. (A) of the present invention for achieving said purpose aspects of the one number (RNA-a guided nuclease) have been introduced new the [ley which will grow oh RNA - guide, said guide including a guide (guide RNA) RNA encoding RNA oligonucleotides including bio-sequence listing and desired target bio-sequence listing is obtained from a library cells using performing DNA sequence analysis; and (b) said guide from the data obtained through sequencing of target sequence pair (pair) RNA - is and it is burnt (indel frequency) including a frequency detecting, evaluating the activity of RNA - guide new the [ley which will grow oh number (RNA-a guided nuclease) method are disclosed. The present invention is a "guide RNA - target sequence pair (pair) library analysis" victims of the method forms a transparent conductive layer, this guide RNA coding is introduced into cells using new the [ley which will grow oh bio-sequence listing and target bio-sequence listing form pair number evaluating activity of RNA - guide means other library method. In particular, the present invention of the present invention to improve purity and yield new the [ley which will grow oh RNA - number determined by means of the activity of the victims of the paired seal has inherent anti-tumor gene in a cell number guide acting on high when congestion exists to confirm the activity of RNA - guide number, the number of active evaluation method of the present invention RNA - new the [ley which will grow oh guide The dummy dielectric calibration/gene, including target-oriented transitions human cells can be incorporated into the animal cell dielectric bio-sequence listing wherein as a technique, animals bearing a specific gene (knock-a out) or brand (knock-a in) or, non-coding DNA sequences - even transitions to produce proteins can be incorporated into the disclosed. The calibration/gene of the present invention method used in the dummy target and non-target RNA - guide new the [ley which will grow oh said dielectric number in which high throughput activity can, this is only used to target location specific operating new the [ley which will grow oh RNA - guide number can be useful. In the present invention the terms "new the [ley which will grow oh guide RNA - number" is used as the desired dielectric anisotropic recognizes the new the [ley which will grow oh number can be, in particular guide RNA of the target by having specificity number new the [ley which will grow oh speak other. The number is the number one new the [ley which will grow oh guide said RNA - but not limited to, specifically microorganism CRISPR Cas9 protein derived from the immune system, specifically Cas9 Cpf1 (CRISPR-a Associated Protein 9) and can be like. Animal including humans cells of said RNA - guide new the [ley which will grow oh number specified in dielectric bio-sequence listing can be again causing duplex cutting (double strand break, DSB), nick (nickase activity) can be formed. (Blunt end) or by cutting of duplex DNA duplex said severance of the two amino (cohesive end) to prevent the adhesive end including both substrate. Homologous recombination in cells is re-joined DSB (homologous recombination) or non-homologous (non non-homologous end-a joining, NHEJ) efficiently repair mechanisms by which a desired target location can be introducing mutations study process. Said RNA - new the [ley which will grow oh guide number is an artificial, or engineered cytolytic resulting (non-a naturally occurring) may be disclosed. Terms in the present invention, the major protein construct element "Cas protein" CRISPR/Cas system, activated to new the [ley which will grow oh [ni car number or the number (nickase) applicable to protein are disclosed. Said Cas crRNA protein is capable of forming a complex (trans-a activating crRNA) (CRISPR RNA) and tracrRNA on order activity can exhibit. NCBI GenBank (National Center for Biotechnology Information) of Cas protein or gene such as publicly known database of information can be achieved. Specifically, said Cas Cas9 protein can be protein. In addition, the Cas protein is the number one but not limited to, Streptococcus ( In the present invention the terms "Cpf1" said CRISPR/Cas system different than the number of new CRISPR as new the [ley which will grow oh, Cpf1 of gene has been reported (Cell, 2015, 163 (3): 759 - 71) makes from oh! acts as scissors. Said Cpf1 number driven by single RNA into new the [ley which will grow oh, is connected to the disk so that a tracrRNA Cas9 has a stage is suitable. In addition, thymine (thymine) PAM (protospacer provided adjacent motif) sequences of DNA rich and a double end (cohesive end) by cutting pressure and to prevent the known as disclosed. Said Cpf1 also relates the number one but not limited to, green onion tax pak in particular candy multi [thwu ( The terms "recombinant" said, e.g. cells, nucleic acid, protein or vectors used that Vref when, introduction of wild-type (native) or heterologous (heterologous) nucleic acid or protein modification of nucleic acid or protein, or modified by cells altered cells are derived from cells, nucleic acid, protein, exhibits or vectors. The, e.g., recombinant Cas9 Cpf1 the proteins include the human codon table (human codon table) or recombinant protein using Cpf1 Cas9 or sequences encoding can be made by reconfiguring. Said Cas9 Cpf1 protein is a nuclear protein or can act in said form may be, cells can be introduced into are easy to form. Protein peptide or protein delivery domain (protein transduction domain) and vertically moving the Cas9 Cpf1 or cells can be connected. Said poly - arginine or HIV TAT protein can be a protein delivery domains may be derived from, the number is not one. In addition said domain peptide or protein delivery cells described example contrast since various kinds is publicly known, various examples the present invention applied to a one skilled said number not one example can. In addition, nucleic acids encoding said Cas9 or Cpf1 protein (nuclear localization signal, NLS) sequence further includes a nuclear position signal can be. The, said Cas9 Cpf1 protein or nucleic acids encoding for the expression of regulatory sequences such as promoter sequences for said Cas9 or Cpf1 including expression cassette includes in addition can be NLS sequence. However the number is not one. Cas9 Cpf1 protein is advantageously connected to separation and/or positive number or tag can be. For example, His tag, Flag tag, such as tag S small peptide tag, or GST (Glutathione S-a transferase) tag, it can be connected along the MBP tag like (Maltose binding protein), the number is not one. The present invention refers to said RNA - guide a number of new the [ley which will grow oh unified method number [...] substrate. Each stage of the detailed hereinafter said method with respect to each other. On the other hand, as the definition of the above discussed said camshafts are represented by terms and aspects a special disclosed. (A) step (guide RNA) coding bio-sequence listing and target RNA oligonucleotides bio-sequence listing including a guide separated DNA obtained from library including cells using performing as deep sequencing (deep sequencing), acted upon by a wide variety of guide RNA target sequence number RNA - guide new the [ley which will grow oh and target and non-target activity is and it is burnt (indel; insertion and deletion) various obtain necessary data analysis generated from cell populations are disclosed. Specifically, step (a), (I) guide RNA coding bio-sequence listing and target bio-sequence listing, i.e. guide RNA oligonucleotide library including a pair (pair) bio-sequence listing and target step number bath, (Ii) said oligonucleotide to improve purity and yield a vector library, in particular step number bath library virus vector, specifically said oligonucleotide library for vector number vector library number each oligonucleotides high pressure liquid coolant in the tank, (Iii) said library of vectors, in particular the cell number in the bath to improve purity and yield virus vector library, specifically, each vector of library of vectors is introduced into said cell, cells involves establishing a library, and (Iv) obtained from a library cells using said DNA sequence analysis, the processing advances to e.g. deep sequencing (deep sequencing) can be performed. (Iv) said step (iii) to obtain said DNA library cells in the cell library established new the [ley which will grow oh number is introduced into the RNA - guide, said culturing cells activity of RNA - guide new the [ley which will grow oh number can be derived. In the present invention is a different property using the terms "library" means the material of at least one homogeneous population (pool or population) 2 included therein. , the other 2 oligonucleotide library bio-sequence listing at least one oligonucleotide, for example guide RNA, PAM sequence, and/or target sequence including the oligonucleotide probes may be different 2 species population, library of vectors (e.g., viral vector library) sequence variation or components including at least one vector population reduction 2 may be, for example, oligonucleotide library vectors in groups of said each for oligonucleotides, corresponding vectors is 2 or more vectors can be a population of oligonucleotides that difference. Cell library 2 having a different property using one or more cells, specifically the purpose of the invention is including other oligonucleotides on each cell can be, e.g. of the number and/or kinds of introduced vector, in particular a population of cells can be different types. In the present invention new the [ley which will grow oh activity of library cells using RNA - guide number in high-throughput mode (high non-throughput manner) is evaluating an incoming data packet, each library constituting said oligonucleotide, vector (e.g., viral vector) and cell of the type can be at least 2 or more, said evaluation method thereof is not one normally operating upper limit number. In the present invention comprises male terms "oligonucleotide (oligonucleotide)" which is connected to the plurality phosphor diester oligonucleotides material it is, the purpose of the invention said duplex DNA oligonucleotide on implementation being. The oligonucleotide sequences used in the present invention said 20 to 300 bp, specifically, 50 to 200 bp, more specifically, may have a length of 100 to 180 bp. In the present invention said oligonucleotides are provided which guide RNA coding comprises bio-sequence listing and target bio-sequence listing. In addition, the oligonucleotide sequences to said PCR can be amplified in the polyester resin composition has a further sequence can be combined can be. Specifically single oligonucleotide RNA target adjacent no Cis provided acting to guide bio-sequence listing can. I.e., in order to identify whether adjacent said guide RNA target bio-sequence listing is for cutting can be designed. said oligonucleotides can be integrated in the chromosome (integration) be a is introduced into the cells. In the present invention terms specific RNA target DNA which is "guide RNA (guide RNA)" which means, all or part of the head is larger RNA - target sequence complementary target sequences can be cutting new the [ley which will grow oh guide number. Typically guide RNA is formed by two RNA, i.e., crRNA tracrRNA (trans-a activating crRNA) (CRISPR RNA) and composed of an RNA including double (dual RNA); or target DNA sequence in sequence and including number 1 site and interacts with the complementary RNA - all or part number sequences including number 2 site or speak form including new the [ley which will grow oh guide, if the target sequence number is new the [ley which will grow oh RNA - guide of the present invention may have an activity on the form without number can be included. In one embodiment, said guide is a guide RNA may be crRNA RNA Cpf1 suppressed, Cas, be applied to a double RNA including in particular Cas9 if tracrRNA crRNA and composed of an single - chain guide RNA fusion form an important part of a form or crRNA and tracrRNA (single-a chain guide RNA; sgRNA) may be in the form disclosed. Target DNA sequence in sequence and said sgRNA is complementary with a portion (Spacer region same, Target DNA recognition sequence, such as base pairing region even cloning) and Cas, in particular Cas9 protein binding (hairpin) structure for hair can be. More specifically, in sequence and complementary target DNA sequence all or part of the portion having an, Cas, in particular Cas9 pin structure and terminator (Terminator) sequence for protein binding can be. The structure described the 5 'in 3' sequentially order can be present. However, the number is not one which, all or part of said guide portion including a major part of the target DNA or RNA is crRNA complementary RNA can be also used in the present invention is cured by any type of guide. Said guide RNA, specifically crRNA sgRNA is complementary in sequence and target DNA sequence or all or a portion of which, or sgRNA crRNA upstream site, specifically sgRNA or crRNA of 5' end that includes at least one additional can. Said additional polynucleotides may be guanine (guanine, G) handler, the one number are not disclosed. In addition, said guide being attached new the [ley which will grow oh RNA is RNA - guide can help scaffolds sequence number. Terms in the present invention, "target bio-sequence listing or target sequence (target sequence)" is new the [ley which will grow oh bio-sequence listing which are expected to target RNA - guide number in either direction a it is, in the present invention of the present invention target bio-sequence listing pair (pair) library analysis method further guide RNA - even sequences comprising the meaning of the analysis through to. In the present invention said oligonucleotide library and includes an RNA target sequence pair on each oligonucleotide library of vectors (pair) form guide and vectors in order, an oligonucleotide or vector RNA corresponding to present guide has a curved target sequence are disclosed. In the present invention active (on a non-target activity) target/non-target activity (or effect, on a non-target effect) (off-a target activity) (or effect, off-a target effect) and said target bio-sequence listing are totally separate sense should it will. All or part of said sequence completely complementary sequences "target active" guide for RNA, RNA - guide is new the [ley which will grow oh cutting sequence number, said cleavage site may cause big dell which is to further activity. "Non-target active" guide RNA sequence completely complementary sequences out some but not all or part of sequences in sequence (mismatch), cutting sequence number is new the [ley which will grow oh RNA - guide, said cleavage site may cause big dell which is to further activity. I.e., non-target activity number sequence that is cut by said target activity and RNA - guide new the [ley which will grow oh RNA sequence complementary to all or part of that guides and completely determined by whether the general outline are disclosed. While, in the present invention "target sequence (target sequence)" is used in the form of pair generated by a new the [ley which will grow oh activity of RNA - guide RNA guide sequence number to determine whether or not action power to each said substrate. I.e., oligonucleotides of the present invention is oligonucleotide library is set can be determined by design (design) or number bath embodiment in that, in said embodiment embodiment includes design phase for target and non-target activity sequences according to the set object pair guide RNA sequences expected activity expected target sequence can be selected and the trust. Said target sequence is RNA - PAM-person (protospacer provided adjacent motif) sequence number new the [ley which will grow oh guide but, the number is not one. The design of oligonucleotides for evaluating the activity of RNA - guide new the [ley which will grow oh number to one skilled can be desired is to perform. For example, aminopeptidase activity specific guide RNA target sequence can be paired to and, in addition said guide RNA target activity or paired non-aminopeptidase sequence can be disclosed. For example, guide RNA sequences, specifically, crRNA sequence completely complementary sequences or some base mismatch some complementary sequences can be designed. In addition, one skilled guide of the present invention the target sequence pair library analysis of oligonucleotides to RNA - additional components may be included in the disclosed. For example, oligonucleotide said direct repetitive sequences, poly T sequence, bar code sequence, invariant portion sequence, promoter sequences, and scaffold further comprises a nucleotide sequence selected from the group consisting but more, the number is not one. Said holes are formed on the oligonucleotides such as length, specifically 100 to 200 can be a made of a bio-sequence listing, the number and not one, kind of RNA - guide new the [ley which will grow oh number used, analysis by one skilled can be appropriately controlled according to the object. On the other hand, the above-described oligonucleotide sequences, 5 'in 3' and guide RNA target sequence can be in the order of the coding sequence, on the contrary 5 'in 3' order guide RNA target sequence and can be designed. For example, said oligonucleotide target sequence, which guide RNA coding sequence, specifically target sequence, bar code sequences, and guide RNA which coding sequence, the following order but can be established, and pass it on one number that are not disclosed. Said oligonucleotide sequences 5 'in 3' order guide RNA coding sequences, bar code sequences, and target sequence can be, specifically guide RNA coding sequences, bar code sequence, PAM sequence, and target sequence can be, guide RNA coding sequences, bar code sequences, target sequence, and PAM can be sequence, guide RNA coding sequences, poly T sequence, bar code sequence, PAM sequence, and target sequence can be, guide RNA coding sequences, poly T sequence, bar code sequences, can be target sequence and PAM sequence. More specifically, direct repetitive sequences, guide RNA coding sequences, bar code sequence, PAM sequence, and target sequence can be, direct repetitive sequences, guide RNA coding sequences, bar code sequences, target sequence, and PAM can be sequence, direct repetitive sequences, guide RNA coding sequences, bar code sequence, PAM sequences, target sequence, and invariant part can be sequence, direct repetitive sequences, guide RNA coding sequences, bar code sequence, target sequence, PAM sequence, and invariant part sequence but, not one number and pass it on. In addition, guide RNA coding sequence number is new the [ley which will grow oh binding RNA - adjacent to guide further comprises a sequence scaffolds assist can be. For example, scaffold sequence, guide RNA coding sequences, bar code sequence, PAM sequence, and can be target sequence. However, pass it on one number are not disclosed. In addition, expression for 5' end site including the promoter sequence can be. In the embodiment of the present disclosure as well as the U6 promoter. 5 'In 3' target sequence order, bar code sequences, and guide RNA can be a coding sequence, specifically target sequence, PAM sequence, bar code sequences, and guide RNA can be a coding sequence, PAM sequences, target sequence, bar code sequences, and guide RNA can be a coding sequence, target sequence, PAM sequence, bar code sequences, poly T sequence, and guide RNA can be a coding sequence, PAM sequence, target sequence, bar code sequences, poly T sequence, and guide RNA can be a coding sequence, than specifically target sequence, PAM sequence, bar code sequence, guide RNA coding sequences, and direct repeat sequences can be, PAM sequences, target sequence, bar code sequence, guide RNA coding sequences, and direct repeat sequences can be, target sequence, PAM sequence, bar code sequences, poly T sequence, guide RNA coding sequences, and direct sequence can be, PAM sequences, target sequence, bar code sequences, poly T sequence, guide RNA coding sequences, and direct repeat sequences can be, invariant part sequences, target sequence, PAM sequence, bar code sequences, poly T sequence, guide RNA coding sequences, and direct repeat sequences can be, invariant part sequences, PAM sequences, target sequence, bar code sequences, poly T sequence, guide RNA coding sequences, and direct repeat sequence but, not one number and pass it on. In addition, guide RNA coding sequence number is new the [ley which will grow oh binding RNA - adjacent to guide further comprises a sequence scaffolds assist can be. For example, target sequence, PAM sequence, bar code sequence, guide RNA coding sequences, and scaffold sequence but, and pass it on one number are not disclosed. In addition, expression for 5' end site including the promoter sequence can be. In addition, as described above, in addition to the aforementioned components said oligonucleotide 5 'and 3' PCR primer attachment sequence further comprises inner end to can be. However, pass it on one number are not disclosed. Said target sequence is 10 to 100 bp, specifically 20 to 50 bp, but there is the length of 23 more specifically than to 34 bp, and pass it on one number are not disclosed. In addition, said guide RNA coding sequence is 10 to 100 bp, specifically 15 to 50 bp, but there is the length of 20 to 30 bp than specifically, and pass it on one number are not disclosed. In addition, each oligonucleotides for nucleotide sequences to identify said bar code sequence is big. Herein said bar code sequence is repeated 2 or more nucleotide (AA, TT, CC, GG) but does not contain a touch-screen, each oligonucleotides designed to identify specifically the number one if not disclosed. In are a plurality of oligonucleotides, each oligonucleotide at least 2 of said bar code sequences may be identified to be a nozzle base is designed. Said bar code sequences may have a length of 5 to 50 bp but, not one number and pass it on. In the embodiment of the present invention in one specific In addition, in the embodiment of the present invention in other Next, said oligonucleotide to improve purity and yield a high pressure liquid coolant (e.g., viral vector) can be a vector library number. Target sequence pair (pair) of the present invention guide RNA - surface of the one of the active evaluation method using RNA - guide new the [ley which will grow oh number using said second paired virus is intended to 30 to 60 seconds. Guide RNA target sequence corresponding to form cells is pair distilled water oligonucleotide library, library of vectors and cell library can be generated in minimizing impact by deviation copy number can be, virus DNA (integration) inserted through the dielectric with time according to target and non-target activity analysis-by temporal expression method and focuses that is, further public welfare (epigenetics) due to the effect of therapeutic factors be relatively's. When said vector is virus, virus library then into cells, can be obtained by producing therefrom, can be a cell infected by using, this procedure using a publicly known method can be properly one skilled performing contrast. In the present invention vector RNA coding bio-sequence listing and target can be oligonucleotides including a respective guide bio-sequence listing. Said vector may be a viral vector or plasmid vector, viral vector or retroviral vectors but specifically lentiviral vectors can be used, the number is not one that are publicly known and purpose of the invention the present one skilled achieved using vector can be freely. Said vector to said intracellular oligonucleotides capable of delivering mediator, e.g. genetic small number big water. Specifically, in the presence of a cell of an individual vector said insert, i.e. oligonucleotides may be developed to insert integral regulation element can be operatively connected. Said vector recombinant DNA techniques to standard number bath and can be positive number. Said vector of the type such as functional in the desired cells to the secretion and eukaryotic cells, particularly not limited. Vector promoter, disclosure codon, a stop codon can and terminator. In addition signal peptide encoding DNA, and/or enhancer sequence, and/or of a desired gene 5 'side and 3' modifications region side, and/or selection marker region, and/or possible prevention unit suitably includes a number like disapproval. In the embodiment of the present invention specific oligonucleotide library in said lentiviral vectors each oligonucleotide performing cloning of lentiviral vector library number (also 4 and 36) have high pressure liquid coolant, obtained virus expression in them. A next step includes, said vector is introduced into the target cell in the cell library number tank are disclosed. Specifically, the method for delivering said high pressure liquid coolant library vector number cells contrast can be achieved using various publicly known method. For example, calcium phosphate - DNA strategy selected, DEAE - dextran - mediated trans [pheyk the law, poly branches - mediated transfection method, electric impact method, fine scanning method, liposome fusion method, a number of method such as fusion of interest into liposomes [pheyk it burns it pushed and can be performed by publicly known it was found. In addition, when using a virus vector, by means of a virus infection (infection) particles object, i.e. vector capable of transferred into cells. In addition, vector can be introduced into cells by gene bambara [...]. Introducing said vector or vector in cells that present itself, but may be integrated into a chromosome, particularly the one number are not disclosed. Herein including oligonucleotides have been introduced guide RNA - library cells prepared by the number target sequence populations of cells said substrate. Each of the cells may be vector, specifically virus types and/or refinish and devices that may be. But, analysis method of the present invention performed using whole cell library and, guide RNA encoding bio-sequence listing and target sequences introduced into cells infection efficiency since form pairs, the deviation of water or copy of oligonucleotides way without being depending on data analysis process from each pair (also 6 to 12 also). Constructing said library to derive new the [ley which will grow oh cells can be introduced into the RNA - guide number dell which is added. Said number is new the [ley which will grow oh introduced cells are types and/or a target sequence having guide RNA - n can be appears to be active according to different degree. Said RNA - or plasmid vector or virus vector number is new the [ley which will grow oh guide cells via delivered, number protein RNA - guide new the [ley which will grow oh itself can be transferred cells, RNA - guide number in a cell can be one that is particularly introducing method number is new the [ley which will grow oh activity thereof is not. For example, protein delivery domain connected form number (e.g., Cas protein, protein Cpf1) RNA - guide new the [ley which will grow oh but such as may be communicated, the one number are not disclosed. Various types can be used publicly known contrast delivery protein domain, as said poly - arginine or derived from HIV TAT protein is cited. However, pass it on one number are not disclosed. In addition, the kind of cells that can said vectors have been introduced, vector types and/or appropriately depending on the type of desired cells to choose one skilled but, for example, Escherichia coli, Streptomyces vaccine compositions which, bacterial cells such as Salmonella mobile [...]; yeast cells; such as Pichia pastoris fungal cells; draw trillion it will bloom,, insect cells such as cells a spotter dope terahertz Sf9; CHO (China Cho cells, chinese hamster ovary cells), SP2/0 (mouse myeloma cells), human (human lymphoblastoid) methods operating, COS, NSO (mouse myeloma cells), 293T, bow melagatran roh e cells, HT-a 1080, BHK (baby hamster kidney cells, baby hamster kidney cells), HEK (human embryonic kidney cells, human embryonic kidney cells), PERC. 6 (Human retinal cells) such as animal cells; or plant cells can be. In said cell library introduced new the [ley which will grow oh new the [ley which will grow oh by number active target sequence pair oligonucleotide and guide RNA - RNA - guide number can be exhibited in a disclosed. I.e., target sequence number for new the [ley which will grow oh introduced by RNA - guide can be DNA scission occurs, the is and it is burnt (indel) can be shown. In the present invention the terms "is and it is burnt (indel)" DNA base sequence is received intermediate (insertion) or deleted (deletion) transition in some base is the general term to each other. As above-mentioned number of duplex DNA RNA - dell which is new the [ley which will grow oh homologous recombination (homologous recombination) or when cutting guide is re-joined by non-homologous (non non-homologous end-a joining, NHEJ) can be introduced into the target sequence in the process where the repair mechanisms. In addition, the activity of DNA sequences of the present invention method is introduced new the [ley which will grow oh RNA - guide number can be obtained from the drops. The nitrile obtained publicly known method such DNA DNA separation can be performed using a variety. Cell library is set cells may be introduced in the base of target sequence occurred in the target sequence is and it is burnt this expected sequence analysis, e.g. deep sequencing (deep sequencing), can be performed according to the obtained data or RNA provided seq. Assay using a method of the present invention target sequence pair library RNA - guide to methods ( The, step (b) said guide from the data obtained through sequencing RNA - n obtain target sequence is and it is burnt frequency (indel frequency) are disclosed. A solder ball is dependent on a target sequence pair (pair) and second guide RNA - dell which is can be generated, the target sequence is and it is burnt by the activity of RNA - new the [ley which will grow oh extent number pairs contained in the guide RNA - said guide can be evaluated. Each pair the oligonucleotide library is set same oligonucleotides can be distinguish specific sequences can be distinguished by inserting a reference data is data method of processing the classification process from distinct sequences analyzed. In one embodiment, the oligonucleotides in the present invention 2 or more nucleotide (i.e., AA, CC, TT, GG) each comprising repeated without, of at least 2 bar code (barcode) sequence each other designed different base included small number-gate. The catabolic pathway in vivo activity of the gene of the present invention acting on paired library high relevance by having new the [ley which will grow oh RNA - guide number, a number of highly active evaluation method more accurately predictable number RNA - guide new the [ley which will grow oh [...] substrate. In the embodiment of the present invention specific gene in a cell number determined by means of the active gene library in the cutting chamber acting on the cutting activity connected with high inherent gene has been confirmed. In addition, high accuracy of the present invention pair library has a number of new the [ley which will grow oh RNA - guide can be activity. Specifically, in the embodiment of the present invention obtained using human CD15 gene library of the present invention pair in other specific activity of priority and basing guide RNA publicly known guide RNA activity of human MED1 gene ranking (Nat Biotechnol, 2014, 32:1262 - 1267, Nat Biotechnol, 2016, 34:184 - 191) by comparing the accuracy of paired library was assessed. As a result, human CD15 and human MED1 gene RNA represented correlation factor applied to guide both high sphere only, publicly known guide RNA has been confirmed that the representing the relation between the active priority and is correlated (37 also). In addition, the methods of the present invention obtained using guide RNA library pair activity of RNA target sequence present in direct correlation with the comparison result obtained by analyzing the activity of guide, by making sure that the high Pearson correlation coefficient, number of active evaluation method of the present invention target sequence pair library using RNA - guide new the [ley which will grow oh guide RNA - high accuracy has been confirmed (38 also). For example new the [ley which will grow oh that is analyzed in the present invention the characteristics of said RNA - guide number, (I) number of RNA - guide new the [ley which will grow oh PAM sequence, (Ii) RNA - guide new the [ley which will grow oh number target active (on a non-target activity), or (Iii) active (off-a target activity) can be non-target RNA - guide new the [ley which will grow oh number comprising. Power to each new the [ley which will grow oh number depending on the characteristics of the design of oligonucleotides can be RNA - guide, which consequently said cells being interpreted from the frequency of the resulting compound and obtained through deep common channel redistribution message is and it is burnt therein. In one embodiment, number of PAM's design of oligonucleotides in RNA - guide new the [ley which will grow oh concern target sequence 5' end base sequences having nucleotide sequences and/or PAM various bio-sequence listing and devices and having potential PAM sequences can. The PAM sequence number corresponding RNA - PAM is and it is burnt by analyzing frequency according to new the [ley which will grow oh guide can be of concern. In the embodiment of the present invention in one specific The other example, target active (on a non-target activity) for analyzing characteristics for various design and a corresponding target sequence or guide RNA, RNA - application of new the [ley which will grow oh guide number consists can be analyzed. Thus maximizing information through guide RNA target design effect can be achieved. In the embodiment of the present invention new the [ley which will grow oh guide number in one specific RNA - or inexpensive price, high activity elicited property of positions of guide RNA or, if there is a GC content of target sequence when the target activity elicited property (also 20 to 22 also), specifically through the other one in the embodiment [leyn mote virus in propagation time between the target activity was (also 23 and 24 also) analysis. To other example, non-target active (off-a target activity) elicited property guided for RNA target sequence for part sequences can be designed out between on and to oligonucleotides, the position of target sequence can be specifically switch design. Effects of the mismatch base according to position of target sequence can be through, this design minimizes the effects of non-target RNA guide information can be obtained. In the embodiment of the present invention in one specific region is a standard of target sequence corresponding guide RNA oligonucleotides with mismatch base design, analyzing relationships with non-target effect angular position of target sequence mismatch base in his (also 25 to 33 also). Number properties of the of the present invention target sequence pair library using RNA - said RNA - guide new the [ley which will grow oh guide number is buried in evaluating the activity of RNA - guide new the [ley which will grow oh show one example, the range of the present invention interprets said example number one capable of free. The core techniques of the present invention target sequence to improve purity and yield by over-paired RNA - guide characteristic including cells ( Another aspect of the present invention, said guide including a guide RNA is RNA oligonucleotides encoding the desired target bio-sequence listing bio-sequence listing and including at least one library 2 cells including cells are disclosed. Another aspect of the present invention, guide RNA encoding a desired target oligonucleotides including bio-sequence listing and said guide RNA is separated bio-sequence listing including vector, and vector library are disclosed. Another aspect of the present invention, said guide including a RNA encoding the desired target bio-sequence listing bio-sequence listing and guide RNA is isolated oligonucleotide, and oligonucleotide library are disclosed. Said cell library, vector, library of vectors, oligonucleotides and oligonucleotide library described in respect of the efined. Another aspect of the present invention (a) involves setting a target bio-sequence listing target RNA - guide new the [ley which will grow oh [thing it does, character number; (b) setting said target bio-sequence listing complementary chain with a base forming a gear pair, guide RNA nucleotide sequence encoding design step; (c) said guide RNA oligonucleotide including a designing step bio-sequence listing the purpose and target; and (d) said step (a) to (c) 1 or more times, specifically including 2 or more times including repeating step, said method for constructing a library oligonucleotides are disclosed. In order to design oligonucleotides represents said oligonucleotide library described efined. This, after determining target sequence, sequences of RNA or RNA sequences for the design guide for PAM including sequence is designed such that out of be a target sequence. I.e., in the present invention analyzing both target and non-target active (on a non-target activity) can be active (off-a target activity), guide RNA sequences fully complementary target sequence or a portion out some sequences (mismatch) state can be complementary. The design process for the guide RNA sequence length and/or bio-sequence listing this difference is I may be designed such that several target sequence, one target sequence for several guide RNA sequence length and/or bio-sequence listing this difference is I may be designed such that, said two process consists of the mixed product can be. Said step (c) or (d) further comprises the steps of synthesizing oligonucleotides can be designed. Another aspect of the present invention in PAM (proto-a spacer provided adjacent Motif) sequence CTTA TTTV or adjacent to, target bio-sequence listing complementary chain (complementary strand) form a pair with a base sequence including, separate guide RNA are disclosed. Another aspect of the present invention separated said guide RNA, or nucleic acids encoding the same including, for correcting dielectric composition. Said separated-out guide RNA can be used together with the guide number is new the [ley which will grow oh Cpf1 RNA - protein. Another aspect of the present invention separated said guide RNA, or nucleic acid encoding the same; and including nucleic acids encoding the same protein or Cpf1, system for calibrating a dielectric in mammalian cells are disclosed. Another aspect of the present invention said guide RNA or nucleic acid encoding the same; and Cpf1 protein or nucleic acid encoding the same sequentially or simultaneously introducing isolated mammalian cell including, dielectric correction in an optical method Cpf1 in mammalian cells are disclosed. As above-mentioned in the present invention Cpf1 protein sequences known in the art TTTN TTTV CTTA or not about proving the PAM-shaped conductor, the present invention through a guide RNA sequence CTTA TTTV or PAM dielectric correction can be useful. RNA - guide of the present invention target sequence pair (pair) library using RNA - number evaluating activity of new the [ley which will grow oh guide to methods method ( Figure 1 shows a Cpf1 also for evaluating the activity of, or paired (pair) indicating a mimetic of oligonucleotides including target sequence and guide RNA sequences are disclosed. Figure 2 shows a map indicating a mimetic also AsCpf 1 lentiviral vectors are disclosed. Psi, packaging signal; RRE, rev response element, WPRE, posttranscriptional regulatory element of woodchuck hepatitis virus; U6, U6 pol III promoter; cPPT, central polypurine tract; EFS, elongation factor 1a short promoter; BlastR, blasticidin resistance gene. Figure 3 shows a map indicating a mimetic also LbCpf 1 lentiviral vectors are disclosed. Psi, packaging signal; RRE, rev response element, WPRE, posttranscriptional regulatory element of woodchuck hepatitis virus; U6, U6 pol III promoter; cPPT, central polypurine tract; EFS, elongation factor 1a short promoter; BlastR, blasticidin resistance gene. Figure 4 shows a high pressure liquid coolant also plasmid library RNA target sequence number for basis vectors (backbone vector) and a paired (pair) and guide including a mimetic lentiviral vectors modified at indicating sequence are disclosed. Psi, packaging signal; RRE, rev response element; WPRE, posttranscriptional regulatory element of woodchuck hepatitis virus; cPPT, central polypurine tract; DR, direct repeat of Cpf1; GS, guide sequence of guide RNA; T, polyT; B, barcode; TS, target sequence; HS, homology sequence; EF1 α, elongation factor 1 α promoter; PuroR, puromycin resistance gene. Figure 5 shows a pair (pair) of the present invention also indicating briefly the other part of the high throughput assay system using a mimetic library are disclosed. Figure 6 oligonucleotide population, each pair (pair) relative copy number plasmid library and cell library are disclosed. Figure 7 paired copy number plasmid library and cell library contains paired copy number plasmid library each oligonucleotide population and standardized are disclosed. Figure 8 each pair relative copy number plasmid library and cell library oligonucleotide population indicating in order copy number are disclosed. Figure 9 relative copy number plasmid library in order copy number indicating each pair library cells are disclosed. Figure 10 plasmid library and oligonucleotide population evaluated correlation between paired deep sequencing (deep sequencing) contains copy number are disclosed. Figure 11 cell library and oligonucleotide population evaluated correlation between paired deep sequencing (deep sequencing) contains copy number are disclosed. Figure 12 cell library and evaluated correlation between paired copy number plasmid library contains deep sequencing (deep sequencing) are disclosed. Figure 13 AsCpf 1 and LbCpf 1 PAM for visually representing a concern of a mimetic process are disclosed. Figure 14 shows a PAM sequence according to empty indicating potential is and it is burnt AsCpf 1 also are disclosed. As his potential PAM ANNNN sequences provide a sequence. Simply to indicate that, "A" is off disagreement. Figure 15 shows a four-TTTN PAM of aminopeptidase is and it is burnt indicating AsCpf 1 also are disclosed. Error bar standard deviation exhibits. * P < 0. 05, ** P < 0. 01, *** P < 0. 001. Figure 16 shows a potential PAM sequence according to indicating empty is and it is burnt LbCpf 1 also are disclosed. As his potential PAM ANNNN sequences provide a sequence. Simply to indicate that, "A" is off disagreement. Figure 17 shows a four-TTTN PAM of aminopeptidase is and it is burnt indicating LbCpf 1 also are disclosed. Error bar standard deviation exhibits. * In the embodiment hereinafter the present invention through a corresponding business are provided as follows. In the embodiment of the present invention is generally described the present invention is to exemplify these for these in the embodiment are not correct limited range. In the embodiment 1: Cpf1 active evaluation method for evaluating the activity of paired library number bath and In the embodiment 1 - 1: oligonucleotide design High-throughput mode (high non-throughput manner) for evaluating the activity of various guide RNA library for Cpf1 plasmid construct, in CustomArray (Bothell, WA) The catabolic pathway position, and introduction locations of frequency is and it is burnt in order to compare, RNA - coding sequences and target sequence two error - free (error-a free) oligonucleotides including 82 Cellemics, Inc (Seoul, a compensation) in copiers. In addition said forward and reverse primer binding sites in PCR amplification oligonucleotide is the same 27 (sequence number 1) can be used to both ends so that base 22 and each including a base with respect to the sequences of (sequence number 2). And each oligonucleotide to be unique to each oligomer (barcode sequence) was inserted into the base bar code sequence 15 central new the [ley mote [tu which will grow. Said bar code sequence is repeated 2 or more nucleotide (i.e., AA, CC, TT, GG) does not contain a rule to, another at least 2 different front and rear of3 all bar code. Each oligonucleotide RNA target sequence (upstream) on each guide bar code sequence located upstream of and downstream with respect to the (downstream). In the embodiment 1 - 2: vector cloning Cpf1 number lentiviral vectors for expressing high pressure liquid coolant, and LbCpf 1 AsCpf 1 encoding plasmid (Addgene; #69982, #69988) derived from plasmid (Addgene; #52962) have sequence number into each lentiCas9 provided Blast prevention, each them (sequence number 3) (sequence number 4) on AsCpf1 Lenti_ provided Blast LbCpf1 Lenti_ provided Blast referred to his (also 2 and 3 also). In addition, plasmid library number for basis vectors to obtain high pressure liquid coolant (backbone vector), a stand-alone number SpCas 9 scaffold region (scaffold region) (Addgene; #52963) lentiGuide provided Puro vector from fluorescence, the vectors are referred to his (sequence number 5) Lenti_gRNA provided Puro vector (4 also). In the embodiment 1 - 3: number of plasmid library bath Number plasmid library for high pressure liquid coolant, said in the embodiment 1 (NEB) oligonucleotide (122 - 130 each base) as Phusion polymer synthesized in a number into a PCR using have, conducting positive number process allowing the gel using MEGAquick provided spinTM Total Fragment DNA Purification Kit (Intron). Then NEBuidler HiFi DNA Assembly Kit (NEB) using the PCR product was assembling said vector Lenti-a gRNA-a Puro positive number. After assembly, said micro l MicroPulser (BioRad) by reaction of 2 with former asthma method with respect to the transformed cells (electrocompetent cells, Lucigen) of 25 micro l electrosurgically through water-insoluble. Then 100 micro g/ml [aym the phosphorus which will bloom LB agar medium containing said transformed cells seeded onto it. Finally library where the number of colonies was secure range of 30 times. Plasmid Maxiprep kit (Qiagen) taken to a plasmid DNA extracted from the colonies. In the embodiment 1 - 4: lentiviral production HEK293T cells (ATCC) to 0. 01% In 100 mm culture dish coated with poly - L - lysine (Sigma) (confluency) him as until 80 - 90% level. In high pressure liquid coolant on carrier number plasmid (transfer plasmid) pMD2 psPAX 2 and said in the embodiment 3. G 4:3 weight ratio: 1 is mixed into. Then, in 100 mm culture dish (Intron Biotechnology) formed in the triangular beam using reagent injection transformed iN non-fect number said plasmid mixture (18 micro g) is intended to him. 15 Transformed implantation has been completed, 12 ml of growth medium was replaced. Said transformation (=15 + 24) and 63 (=15 + 48) time after virus injection from 39 including collection of supernatant. Virus-free medium containing 1 - 2 difference (batch) tea and disposed mixing, 4 °C 5 minutes was 3,000 rpm in cyclone. Then, Millex-a HV 0. 45 Micro m (low protein binding membrane, Millipore) was filtered supernatant that protein combination just to use after said -80 °C until his of the installed application. In the embodiment 1 - 5: generation of library cells To make the library cells, 100 mm dish attached to lentiviral vectors HEK293T cells (1. 5 X 106 - 2. 0 X 106 ) Transduced to him. After transduction from 3, 2 of 3 to 5 g/ml was during the [phyu it is rome micro cells. The study of the present invention to preserve blocks library, said library containing a 3 x 10 to minimize the6 /100 Mm culture dish to a density of two hardships. In prevention of endogenous human gene number number WPRE lentiviral vector element In the embodiment 1 - 6: Cpf1 delivery to library cells AsCpf 1 or LbCpf1 - expression of recombinant lentivirus vectors for transduction of, first cell library (2 x 106 - 3 X 106 Two) 100 mm culture dish was seeded onto 24 time of the transfection. In DMEM 10% FBS (fetal bovine serum, Gibco) then containing AsCpf1 - expression virus vector transduced cells after, 10 micro g/ml in DMEM containing 10% FBS and blinder [...] S (blasticidin S, InvivoGen) hardships. AsCpf 1 or LbCpf1 - coding plasmid DNA sequence in the case of implantation, said first cell library (3 x 106 Two) was 60 mm dish of transfecting injection 6 time of the inoculating 3. Then plasmid LbCpf1 Lenti_ provided Blast AsCpf1 Lenti_ provided Blast or micro g 4, 2000 (Invitrogen) liposomes to cells injection with respect to the micro l [pheyk it burns it pushed and 8. After the queue hemp cloth which is [syen grudge night, DMEM medium containing 10% FBS to replace him. Transformed cells after administration of 10 micro g/mL then injected from a culture medium containing 4 [...] blinder in him as during the third 1. In the embodiment 1 - 7: deep sequencing (deep sequencing) Wizard Genomic DNA purification kit (Promega) was separating dielectric from a library cells using DNA. Then Phusion (NEB) embedded using frequency analysis is and it is burnt compensated with respect to the PCR amplified target sequence produced polymer. 100 Times in order to achieve a range of cells of the library (coverage), PCR DNA was used per sample to the mold at a micro g dielectric 13 1 difference (1 x 106 10 Micro g of 293T cells assumed a dielectric DNA). Each samples, each reaction 1 micro g of DNA per 50 micro l have independent Conference 13 dielectric to performing an electrochemical reaction, reaction-product is mixed. In order to compare a frequency position and introduction locations of the catabolic pathway is and it is burnt, 100 ng of DNA per sample target sequence amplification target sequence using PCR - been been introduced and the catabolic pathway. (Intron) was then PCR product using a positive number MEGAquick provided spinTM Total Fragment DNA Purification Kit. 2 PCR PCR product obtained in a heating element adapter 20 ng illumination and switching said 1 difference number difference (Illumina adaptor) and bar code sequences with is configured to receive. PCR reactions at the table 1 antibody represented to have shown. Separating the final end product, positive number, after mixing was analyzed using MiSeq or HiSeq (Illumina). FP: forward primer (forward primer), RP: reverse primer (reverse primer). In the embodiment 1 - 8: pair copy number (Pair copy number) analysis To find a copy of each pair in library number, read the next formula with the number stored in the standard. In the embodiment 1 - 9: is and it is burnt frequency analysis Using custom fine line SCRIPT (custom Python scripts) classification was detected via a cooling sequencing data analysis. 15 - Oxidase (constant sequence) 4 - base invariant portion downstream of base bar code sequences, i.e. sequences of data classification based on total 19 - base pair (pair) in a respective guide target RNA - by dielectrophoresis. Expected cut position (i.e., cut position intermediate 8 bp region) located around the Cpf1 inserts or deletions caused by mutations was not considered. Number been [...] single base the substitution is in assays. Cpf1 activity of RNA derived from a chamber contained in the guide number is and it is burnt, is and it is burnt observed in cells semiconductor integrated background subtracted frequency is and it is burnt Cpf1 frequency transmitting the bill. Said frequency background is and it is burnt mainly generated in oligonucleotide synthesis. In order to enhance the accuracy of analysis, the classification number read background is and it is burnt (table 2) was deep sequencing data according to use frequency per pair. In the embodiment 1 - 10: comparison of is and it is burnt frequency HEK293T cells 48 well dish to form a guide RNA - coding sequences and target sequence including lentiviral vector was transduced independent. 3 After transfection, transduction cells and the non-number for a stand-alone, a leucine (puromycin) was 3 to 5 2 μg/mL [phyu rome said liver cells. Said transduction cells expressing lentiviral vector Cpf1 AsCpf 1 as above-mentioned was transmitting. Cpf1 after introducing 5, conducting a DNA deep separated from the common channel redistribution message. In the embodiment 1 - 11: computation of chromatin accessibility Once the presence of chromosome 17 and 22 times that number and [...] each copy cells 4, 4 from the dielectric of genome region optionally a minor. In the region of the two said 4 (loci) at an arbitrary position on a guide 82 total RNA was design to target. ENCODE (ENCFF000SPE) derived from the data and a DNase I sensitivity score (sensitivity score) DNase provided seq the bill. A target at the position of each said DNase I sensitivity score, the corresponding location in overlapping DNase provided seq sequencing read fragments (DNase provided seq sequencing read fragments) first counted number of the bill. For example, in cases of overlap of two target site (position 5) read sequencing 5 position, said position is acquired his [su nose 2. The length of the joints of each PAM and including 27 bp target sequence are disclosed. The, DNase I sensitivity score target site at the position of each of two score was obtained by average 27. Human genome of 3 billion position 32 (hg19/GRCh37 from UCSC genome browser) 82 of DNase I score when ordered two target site was a wide distribution score (0% - 99. 99%). In the embodiment 2: Cas9 active evaluation method for evaluating the activity of paired library number bath and The present invention is new the [ley which will grow oh victims of the other is injected RNA - guide number, number of active evaluation method of the present invention RNA - guide using new the [ley which will grow oh SpCas 9 has been confirmed. In the embodiment 2 - 1: oligonucleotide design High-throughput mode (high non-throughput manner) for evaluating the activity of various guide RNA library for SpCas 9 plasmid construct, the victims of the in the embodiment the present invention method similar to a guide oligonucleotide target sequence RNA - front and rear. Specifically, CustomArray (Bothell, WA), Twist Bioscience (San Francisco, CA) in In the embodiment 2 - 2: number of plasmid library bath In plasmid library number number work grudge oligonucleotides for said in the embodiment including high pressure liquid coolant, said Phusion (NEB) using PCR oligonucleotide (120 each base) as a polymer into a number have, MEGAquick provided spinTM Total Fragment DNA Purification Kit (Intron) allowing the gel using conducting positive number process. Then NEBuidler HiFi DNA Assembly Kit (NEB) using PCR product was a positive number LentiGuide_Puro (Addgene, #52963) assembling said vector. After assembly, said micro l MicroPulser (BioRad) by reaction of 2 with former asthma method with respect to the transformed cells (electrocompetent cells, Lucigen) of 25 micro l electrosurgically through water-insoluble. Then 100 micro g/ml [aym the phosphorus which will bloom LB agar medium containing said transformed cells seeded onto it. Finally library where the number of colonies was in the range of 17 - 18 times ensure. Plasmid Maxiprep kit (Qiagen) taken to a plasmid DNA extracted from the colonies. In the embodiment 2 - 3: lentiviral production HEK293T cells (ATCC) to 0. 01% In 100 mm culture dish coated with poly - L - lysine (Sigma) (confluency) him as until 80 - 90% level. Number carrier on said in the embodiment 2 - 2 plasmid (transfer plasmid) in high pressure liquid coolant and pMD2 psPAX 2. G 4:3 weight ratio: 1 is mixed into. Then, in 100 mm culture dish (Intron Biotechnology) formed in the triangular beam using reagent injection transformed iN non-fect number said plasmid mixture (18 micro g) is intended to him. 15 Transformed implantation has been completed, 12 ml of growth medium was replaced. Said transformation (=15 + 24) and 63 (=15 + 48) time after virus injection from 39 including collection of supernatant. Virus-free medium containing 1 - 2 difference (batch) tea and disposed mixing, 4 °C 5 minutes was 3,000 rpm in cyclone. Then, Millex-a HV 0. 45 Micro m (low protein binding membrane, Millipore) was filtered supernatant that protein combination just to use after said -80 °C until his of the installed application. In the embodiment 2 - 4: generation of library cells To make the library cells including said oligonucleotides, HEK293T cells attached to first transistor 150 mm dish number lentiviral vectors high pressure liquid coolant in said in the embodiment (7 per each dish. 0 X 106 Two cells) was to be transduced. After transduction from 3, 2 of 3 to 5 g/ml was during the [phyu it is rome micro cells. The study of the present invention to preserve blocks library, said library cells containing three 150 mm dish (7 per each dish. 0 X 106 Two cell density) was held. In the embodiment 2 - 5: Cas9 delivery to library cells SpCas 9 expression lentiviral vectors for transduction of, first number in said in the embodiment trillion one year-old cloth library (2. 1 X 107 Two) was inoculated with a culture dish made in three 150 mm 24 time of the transfection. In DMEM 10% FBS (fetal bovine serum, Gibco) then containing SpCas9 - expression virus vector transduced cells after, 10 micro g/ml in DMEM containing 10% FBS and blinder [...] S (blasticidin S, InvivoGen) hardships. In the embodiment 2 - 6: deep sequencing (deep sequencing) Wizard Genomic DNA purification kit (Promega) was separating cells using high pressure liquid coolant in said in the embodiment number dielectric from a library DNA. Then Phusion (NEB) embedded using frequency analysis is and it is burnt compensated with respect to the PCR amplified target sequence produced polymer. 100 Times in order to achieve a range of cells of the library (coverage), PCR DNA was used per sample to the mold at a micro g dielectric 180 1 difference (1 x 106 10 Micro g of 293T cells assumed a dielectric DNA). Each samples, each reaction 2 micro g of DNA per 50 to 90 micro l have dielectric independent Conference of performing an electrochemical reaction, reaction-product is mixed. (Intron) was then PCR product using a positive number MEGAquick provided spinTM Total Fragment DNA Purification Kit. 2 PCR PCR product obtained in a heating element adapter 20 ng illumination and switching said 1 difference number difference (Illumina adaptor) and bar code sequences with is configured to receive. PCR reactions at the table 3 antibody represented to have shown. Separating the final end product, positive number, after mixing was analyzed using MiSeq or HiSeq (Illumina). The present invention using the victims of the library pairs prepared by the number from said in the embodiment, similar to active evaluation of active evaluation method in said in the embodiment Cpf1 Cas9 by dielectrophoresis. Experiment example 1: pair library using Cpf1 active evaluation of Experimental example 1 - 1: guide RNA - development of target sequence pair (pair) library High-throughput mode (high non-throughput manner) to evaluate the activity of various guide RNA Cpf1 with, the present invention target sequence pair library RNA - victims of the guide number was high pressure liquid coolant. Said guide RNA sequences and a corresponding target sequence including 11,961 - array of a PCR amplified population (pool of array-a synthesized oligonucleotides; also 1) made from synthetic oligonucleotides, [...] assembly using lentiviral plasmid brasiliensis (4 also). Repetitive sequences (sequence number 20) directly connected to the base and has a forward, guide sequences for crRNA polyadenine nucleotide. The PAM target sequence which sequence, invariant part (constant; sequence number 21) connected to the base part has a backward to the attachment location is constant region vector (contant region vector annealing site) are disclosed. Cloned plasmid is the sequence of the sequence number 3 has bio-sequence listing of said via. Guide RNA corresponding cells expressing target sequence to make the library of cells including in dielectric, said number HEK293T cells (also 5) was prepared by the lentiviral library plasmid from a library. Then, inserted guide RNA target sequence by cutting and formation of a dielectric is and it is burnt to, inject or transformed cells expressing plasmid encoding Cpf1 Cpf1 lentiviral vectors to be transduced cells, said cells deliver Cpf1 his library. Then, PCR amplified target sequence and, for evaluating the frequency detected via a cooling sequencing - based analysis of conducting is and it is burnt. As a result, oligonucleotide population through each pair and devices relative copy that deep common channel redistribution message has been confirmed. I.e., copy number, maximum copy protrusion having an upper 0. 5%, And a lower protrusion having minimum copy 0. 5% To 99% in number the oligonucleotide probes [...] copy number has been confirmed that the difference I maximum 130 times the oligonucleotide probes (6 also). Oligonucleotide population as compared to the copy number plasmids and cells in slightly higher levels of deviation her library. The, copy number plasmids and paired oligonucleotide and plasmid copy number to each cell library cells respectively paired regulation, as a result oligonucleotides compared to those occurring in the population of copy number variation has been confirmed that the low level of deviation (also 7). Plasmid library and cell library creation process further generates most of the copy number variation range paired oligonucleotide and plasmid copy number variation caused by each library as signal peptides (also 8 and 9 also). Oligonucleotide population, plasmid library, and cell library copy of each pager number correlation (also 10 to 12 also) was very high. The same classifies, such variation number tank supplying library cells, i.e. [...] assembly, transformed, lentiviral vector number bath, and while such as transduction process is increased, each library cells n copy number variation deviation copy number of oligonucleotides will mainly caused by big. On the other hand, cell library MOI in about 7. 0 Min. Table 4 is a table to organize his oligonucleotide design and analysis filtering conditions are disclosed. The number of oligonucleotides to table 5 is new the [ley which will grow mote [tu cluster and cells in pair portability library table are disclosed. Experiment example 1 - 2: position and introduction locations of the catabolic pathway is and it is burnt frequency comparison The present invention is introduced by victims of the catabolic pathway [leyn in mote virus genome position and a corresponding target specified sequence is and it is burnt in a strong correlation between frequency synthesis in a position that has been confirmed that the found (40 also). Such a high correlation is shown when paired to improve purity and yield higher levels than the uncomplexed analyte. Cas9 - mediated chromatin accessibility is and it is burnt formation efficiency (chromatin accessibility) affecting the catabolic pathway is different but the recessed portion, [leyn mote virus (integrate) introduced in the areas of the transfer case further because, introduced at the sites where the denaturing detergent are expected to provide a high degree of accessibility. Where the difference of the catabolic pathway that chromatin accessibility is and it is burnt in order to reduce a frequency change, the present invention with the victims of the subset of similar chromatin accessibility (subset) is and it is burnt in the catabolic pathway region introduced with regions of the correlation between frequency was compared. To this end, Encyclopedia of DNA element (ENCODE) obtained in HEK293T cells staining using data obtained from DNase I hypersensitivity DNase0seq value calculating his accessibility. As a result, similar chromatin accessibility score target subset having high relevancy in, in particular, high chromatin accessibility has been confirmed that the subset having higher (41 and 42 also). In most target sequence, sequence contained in the target frequency is and it is burnt in the catabolic pathway is and it is burnt introduced in corresponding higher than we shall, in particular higher in his low chromatin accessibility. In addition, cell library copy of each component in the number, similar library used in previous studies (6 to 11 also) variability between themselves. On the other hand, about 7 average MOI library cells. 0 Pulse, strong correlation between two biological simulation body (replicate) numbers. The delivery of two different cell to a library Cpf1 caused frequency was similar is and it is burnt. (43 Also). In addition, two different Cpf1 method (Cpf1 encoding plasmid temporary transfected (transient transfection) and Cpf1 transduction of lentiviral vector encoding (transduction)) when delivered, the victims of the present a clear association has been confirmed that the present invention is and it is burnt frequency (also 44). In most analysis target sequences, is and it is burnt after transduction of lentiviral vector expressing Cpf1 higher frequency (also 44) has been confirmed. , the victims of the present invention, plasmid transfected proceeding the number experiments and determining a temporary LbCpf 1 PAM [...], Cpf1 through transduction of lentiviral vectors provide a of abortion. Experiment example 1 - 3: identification of PAM sequence in a mammalian cell The present invention of the present invention the victims of the AsCpf 1, crRNA, and target DNA of joint decision (co-a crystal) structure, which does not comprise the four PAM th a base, base first cells (5 '- TTT-a 3') is Cpf1 protein exhibits interact, "5 '- TTTN-a 3'" PAM sequence as a supporting each other. In a mammalian cell in vivo studies of the present invention to the preferred TTTN TTTV from user verification for PAM helps understanding. Additionally, the TTTA AsCpf 1 is and it is burnt (not of the non-is and it is burnt LbCpf 1) frequency of PAM is used as sequence when low levels of significantly high has been confirmed. This, as compared to other potential PAM TTTA AsCpf 1 sequence of PAM is finely by preferred for clinical use other. Next, the victims of the TTTA PAM of the present invention 5' end near base affecting efficiency was assessed whether to alter the dielectric calibration. ATTTA, tTTTA, cTTTA, and gTTTA is and it is burnt between frequency change (also 19, 39 and of a) while service is, is and it is burnt cTTTA LbCpf 1 when used in frequency of PAM sequence is has been confirmed that a significant levels less than tTTTA aTTTA or high (of Figure 39 b) Experiment example 1 - 4: target activity (high-a throughput profiling) high throughput profiling The present invention then victims of the guide associated with RNA target sequence function key features of his efficiency. The screening of a plurality of guide RNA when considered essential by dielectric calibration in start step, the dielectric characteristics of identifying modulators of target sequence will promote development of the dummy. The present invention first AsCpf 1 and the victims of the The next most high activity for 20% guide RNA, nucleotide at the position of each of target sequence AsCpf 1 preference a visit from the police. The most features to the immediate vicinity of the nucleotide sequence located a PAM sequence differences, been observed in 1 position. High activity in the gas guide RNA (thymine) thymine bases located at said 1 significantly reduced (21 also). Although sequence-specific features include a different but, even the position of the next said PAM SpCas 9 very important disclosed. The present invention does not he is in the mote it pushed the chips 1 victims of the target protein that bind to number 1 position of Cpf1 effected crRNA new the [ley which will grow oh oh stand, not stabilized and determining the interaction between concerned with him. Based on DNA - binding AsCpf 1 structure (PDB 5B 43), WED hydroxy side chain is within a domain of N2 to form stable Thr16 guanine bases or polar interaction, uracil and mote public opinion form the event is O2 (39 also). However, the hydroxyl side chain interaction with the oscillation moiety Thr16 adenine is not present, the position of adenine ribonucleotides crRNA unstable or disclosed. The, target DNA present in the mote it pushed position of strand 1 does not preferred. Finally, the present invention is 40 - 60% GC content AsCpf 1 between the parties for target sequence has been confirmed that the highest activity (22 also). This SpCas 9 relative to previous result similar also are disclosed. In addition Cas9 is and it is burnt guide has affected by the length of time that frequency expressed in cellular RNA. 6 To 11 after transduction of lentiviral vector expressing Cas9 previous study and guide RNA such as when long-term culture, time-dependent increases in frequency and severity is and it is burnt-out efficiency over time that reported possible disclosed. If only a small number of studies however hi (two 1, 5 or 6) relatively short guide RNA (up 14) since only during testing, in long-term culture guide RNA coding efficiency limits sufficient to overcome frequency is and it is burnt may cause his clearly whether or not. Screening efficiency is and it is burnt frequency is significantly affecting levels of screening in both basic research and new the [ley which will grow oh [...] number (i.e., Cas9) on dielectric guide RNA is because lentiviral vector, this important door number are disclosed. , the present invention is victims of the month (31) about said door number is and it is burnt to analyze the frequency expressed RNA 220 to the guide described his wishes. AsCpf 1 when delivered to a lentiviral vector, 5 increasing culture time is and it is burnt up significantly been increased (also 23 and 24 also) both average and each frequency. This SpCas 9 similar to prior observations are disclosed. However, the difference frequency is and it is burnt after transduction of 5, 10 and 31 cannot. This culture of 5 or more is principally determined by the sequence specific RNA target and guide do not increased over the frequency level is and it is burnt for clinical use are disclosed. Experiment example 1 - 4: non-target activity of high throughput profiling Next, Cpf1 evaluating his intended target of active profile (off-a target activity profile). As the first step, a guide RNA target sequence of mismatch effect high cutting efficiency by identifying (mismatch effect) was intended. The, AsCpf 1 to 4 at the guide on RNA target sequence have corresponding design, these targets for the frequency of transduction is and it is burnt after each 5 53%, 34%, 32%, and 15% phosphorus has been confirmed. One of the 3 highest target cutting efficiency have selected for profiling effect non-target RNA at the guide, said guide RNA target sequence angular position with his bio-sequence listing mismatch effect analysis (also 25). As a result 1 to 6 times in situ, one bp is and it is burnt and to reduce the frequency mismatch is confirmed (26 also). Said seed region (seed region) which means that the result is this position are disclosed. Said of the present invention such as Further, at positions 7 to 18 one bio-sequence listing frequency period is and it is burnt mismatches can been an intermediate level (26 also). The present invention is pressed victims of the trunk region (trunk region) was designated. The present invention results from the victims of the trunk region of nucleotide (nt) RNA AsCpf 1 seed region in guide 18 bio-sequence listing of mismatches can cannot (intolerable) in a highly efficient, highly efficient (tolerable) can miss pro queue earth bio-sequence listing mismatches can in the region is determined to be off. This, of FnCpf 1 Thus, the present invention then victims of cutting guide RNA target and non-target cell through the use effect his analysis. As a result, guide of RNA 3' end up reducing the length to the guide RNA target is and it is burnt 4 nt cutting or minimum 19 nt non-maintaining frequency has been confirmed that the frequency that target is and it is burnt gradually decreased (27 also). This SpCas 9 observed an effect analogous to guide cutting without reduction of target cell through the use effect to reduce the effect of non-target RNA may be big. Experiment example 1 - 5: target location is and it is burnt frequencies having a high degree of correlation of the catabolic pathway Cpf1 activity of library-based assessment The present invention analyzing the victims of the discordance between the number base relevance of his non-target effect. As a result, mismatch in a database of potential non-target location base connected with the non-target effect has been confirmed (28 also). The present invention further victims of the incorporates a region, trunk seed region is followed, trunk region, the trunk and miss pro queue earth between subsequent area, miss pro queue earth region was assessed the effect of a base water mismatch in five. As a result all mismatch in a base water increases as the low frequency is and it is burnt causing recording has been confirmed. But, in the area of the frequency mismatch of 4 or 5 exhibits a significant deep queue earth tends to miss pro is and it is burnt such borderline (29 and 30 also also) appeared. In addition, seed or seed trunk is followed by one or more mismatches can substantially completely is and it is burnt in a region formation was number 3 billion. Next, the present invention is in the form of non-target effect affecting visit from the police whether victims of the mismatch. Seed region and trunk region, non-wobble transition (non-a wobble transition) or divert mismatch (transversion mismatches) that may be associated with high frequency wobble transition mismatch (wobble transition mismatches) than is and it is burnt has been confirmed (31 to 33 may also). This SpCas 9 (unbiased) target for the effects of non-deflection to coincide in analysis results are disclosed. Only, the phenomenon is observed in the area of the frequency mismatch is and it is burnt all types of reducing not only slightly miss pro queue earth. Experiment example 2: pair library using Cas9 active evaluation of Experimental example 2 - 1: Cas9 active evaluation number bath for pair library High-throughput mode (high non-throughput manner) to evaluate the activity of various guide RNA Cas9 with, the present invention target sequence pair library RNA - victims of the guide number was high pressure liquid coolant. Said guide and a corresponding target sequence including 89,592 RNA sequence of a PCR amplified population (pool of array in a non-synthesized oligonucleotides; also 35) made from the array - synthetic oligonucleotides, [...] assembly using lentiviral plasmid brasiliensis (36 also). Guide RNA corresponding cells expressing target sequence to make the library of cells including in dielectric, said number HEK293T cells (also 5) was prepared by the lentiviral library plasmid from a library. Then, inserted guide RNA target sequence by cutting and formation of a dielectric is and it is burnt to, Cas9 lentiviral vectors expressing the transduced cells, said cells deliver Cas9 his library. Then, PCR amplified target sequence and, for evaluating the frequency detected via a cooling sequencing - based analysis of conducting is and it is burnt. Experimental example 2 - 2: human CD15 gene and human MED1 gene RNA Cas9 for active evaluation guide A pair number in said in the embodiment high pressure liquid coolant to improve purity and yield for human CD15 gene and human MED1 gene guide RNA Cas9 activity was assessed. Specifically, paired guide RNA library obtained using active priority and Nat Biotechnol, 2014, 32:1262 - 1267, Nat Biotechnol, 2016, 34:184 - 191 by comparing the activity of RNA library was assessed accuracy of ranking guide nucleotide pairs. As a result, human CD15 gene RNA of each sphere only applied to guide correlation coefficient R=0. 634 Precursor a, human MED1 gene (exon in upper 80% of the overall length of designed) applied to guide RNA of each sphere only correlation coefficient R=0. 582 Represented a, publicly known both active priority and is correlated representing the relation between the two paired guide RNA library has been confirmed (37 also). Experimental example 2 - 3: intracellular target sequence a guide for guide RNA RNA activity and pair library active comparison The present invention is obtained by using guide RNA library pair victims of the activity of a cell and method directly target sequence activity of guide RNA was obtained by analyzing the correlation of a leakage. Specifically, HEK293T cells 48 a-well dish after seeding, including lentiviral vector transduced target sequence was paired guide RNA -. 3 Concentration of 2 μg/ml after the [phyu it is rome transduced cells transduced cells was only selecting processing. In DMEM 10% FBS (fetal bovine serum, Gibco) then containing SpCas9 - expression virus vector transduced cells after, 10 micro g/ml in DMEM containing 10% FBS and blinder [...] S (blasticidin S, InvivoGen) hardships. SpCas9 - 6 Wizard Genomic DNA purification kit (Promega) using cells from a library dielectric DNA expression virus transduction after separating him. (NEB) [leyn with mote virus produced using frequency analysis is and it is burnt then compensated Phusion polymer inserted into a cell target sequence with respect to each PCR amplified target sequence. Each samples, each reaction using 100 ng DNA per reaction of 20 micro l conducting dielectric. (Intron) was then PCR product using a positive number MEGAquick provided spinTM Total Fragment DNA Purification Kit. 2 PCR PCR product obtained in a heating element adapter 20 ng illumination and switching said 1 difference number difference (Illumina adaptor) and bar code sequences with is configured to receive. Table 3 used in PCR reaction to said primer comprises a shown. Separating the final end product, positive number, after mixing was analyzed using MiSeq or HiSeq (Illumina). As a result, intracellular target sequence a guide for guide RNA active Pearson correlation coefficient R=0 RNA activity and pair library. 546 Charge high correlation has been confirmed. Therefrom, of the present invention SpCas 9 guide RNA - using high-throughput mode with high accuracy assessment of the target sequence pair library has been confirmed (38 also). Experiment example 3: active evaluation method in comparison with traditional Cpf1 target sequence Active evaluation method of the present invention high-throughput mode between an individual evaluation method was compared. Specifically, to cost USD, labor time unit is shown below the maximum amount for which human skills. Culture time such as interrupt (break) in the event of greater than time, labor is not calculated. The result shown to table 6. Both the classifies said result, the present invention refers to a particular target sequence in a mammalian cell RNA activity a number identifying a guide for high throughput evaluation method can be know [...]. RNA for specific gene knock-out of calibration or dielectric in particular regions on dielectric can be guide design, the advantage of simple delivery means such as injection (transient transfection) is and it is burnt primarily through temporary frequency are identified. However, efficiency of transformation frequency guide RNA itself is and it is burnt affected as well as the injection efficiency. Thus, the identifying method traits implantation or transfer efficiency variation frequency is and it is burnt optimal guide RNA sequences can be identified by stable cannot. In the present invention and/or a population of cells transformed implantation efficiency of transduction of a single batch in the at least one guide RNA 10,000 once when the confirmation, in another configuration transferred between deviation stored in the error tolerance can be induced. The common case where a somewhat lower traits implantation or transduction efficiency rather than efficiency of all guide RNA tested but which is able to reduce, activity of ranking or "relative" activity guide RNA thus, among tested guide RNA separation process from the highest activity. Other delivery efficiency can be caused by a fog is by convective method conducts an experiment repeated, and the compressed refrigerant is top substrate. Further, the methods of the present invention pair library using method and the status of the diversifies the therapeutic factor not influenced according as public welfare. The lentiviral vector is primarily inserted into transfer active region, lentiviral vector delivery therapeutic state can be a population of cells that pair library public welfare caused is and it is burnt can be compared with a frequency. Transfer efficiency, cells of severe door number comparing RNA efficiency variation state and cells for his number one guide portion which inserted. However, paired RNA sequence library of the present invention can be stably guide based on evaluating efficiency, transmission or public welfare efficiency such as returning a plurality of delay is reduced likelihood that the therapeutic affect. -1,400 Transformed cell by injecting a plasmid encoding two levels of guide RNA - cavity, public welfare and composition containing the therapeutic condition as the guide RNA activity can identify parameters affecting the fair which freezes in the case of levels of middle size (unpair) double library approach, each cell can be expressed in a transgenic plant in which a hollow guide RNA library are implanted to a plurality of non-target effect analysis difficult for, the RNA is and it is burnt this determines whether that is difficult to be identified by any guide has disadvantages. Further, guide RNA copy number to significantly affect cutting efficiency, in this case n semiconductor integrated copy number variation in a respective guide pin is significant difficulty predicting activity of the RNA. Similar to existing known libraries, library of the present invention present even copy number of deviation. However, when of the present invention, guide RNA and target sequence pair (pair) used to form, guide RNA synthesized precedence do not correspond to the target sequence is a linear or cyclic delivered even when several pairs of cellular and ignore comparable levels, substantially all cells and a corresponding target DNA sequence coding the specific guide RNA synthesis associated with the presence of one transforming copy number variations thereof can film. Even non-target evaluation even by a similar target sequence, more than copy number are not introduced or more particular guide RNA sequences, other pair and the reaction between the target sequence do not appear significant level guide RNA target can be assessing the non-driven end. Further, dilution and adjusting the lentiviral vectors can be introduced copy number. The present invention refers to in addition RNA - guide dielectric can be determining a parameter affecting the operation. I.e., target sequence, effector new the [ley which will grow oh kind of number five brush logs (effector nuclease ortholog), guide RNA structural region, therapeutic target DNA of public welfare state, exposing new the [ley which will grow oh effector RNA guide number concentration and period, and guide RNA such as transfer efficiency factor effector new the [ley which will grow oh number is and it is burnt in identifying target and non-target location be frequency percentage which it will do. Each parameter of the present invention via high-throughput mode with various target sequence pair library may be tested for their effectiveness and hif2e.. The classifies said result, the present invention refers to new means for detecting non-target effect number [...] substrate. Non-target effects estimated sequence - guide The number of the studies in the field guide RNA - new the [ley which will grow oh 'industrial revolutionary' can be compared, the number of the existing method was difficult individual measuring method (a cottage system) number RNA - guide new the [ley which will grow oh activity through championship library not based, high-throughput mode (a factory system) to Description of or more from, the present invention of the present invention is provided to the one skilled technical idea or essential characteristics thereof without changing other specific embodiment can form can be understand are disclosed. In this regard, in the embodiment described above are exemplary in all of which was not understood to definitive must substrate. The meaning of the description of the present invention carry patent said range rather than if all of the form of the present invention derived from equivalent general outline and range and method for changing or modified range should interpreted. The present invention relates to a method for evaluating RNA-guided nuclease activity in cells in a high-throughput manner, and specifically, to a method for evaluating RNA-guided nuclease activity from the indel frequency of a cell library, including separated oligonucleotides including coding nucleotide sequences of guide RNA and target nucleotide sequences. According to the present invention, the method for analyzing the characteristics of RNA-guided nucleases using the library of guide RNA-target sequence pairs is an in vivo high-throughput method that allows evaluation of RNA-guided nuclease activity, and therefore, the method can be effectively utilized in all fields where RNA-guided nucleases are applied. COPYRIGHT KIPO 2017 (A) new the [ley which will grow oh RNA - guide number (RNA-a guided nuclease) have been introduced, guide (guide RNA) encoding said RNA oligonucleotides including cells including a desired target bio-sequence listing bio-sequence listing and guide RNA is obtained from a library using performing DNA sequence analysis; and (b) said guide from the data obtained through sequencing RNA - including a target sequence pair (pair) of detecting frequency is and it is burnt (indel frequency), evaluating the activity of RNA - guide new the [ley which will grow oh number (RNA-a guided nuclease) method. According to Claim 1, including said guide RNA is a crRNA (CRISPR RNA) will, method. According to Claim 1, said PAM (protospacer adjacent motif) oligonucleotide sequences including sequences, method. According to Claim 1, the oligonucleotides can be 5 'in 3' or a second, guide RNA coding sequences, bar code sequences, and target bio-sequence listing including a, method. According to Claim 1, the oligonucleotides can be 5 'in 3' or a second, guide RNA coding sequences, bar code sequence, PAM (protospacer adjacent motif) sequence, including a target bio-sequence listing, method. According to one of Claim 1 to Claim 5, said oligonucleotides are provided which direct repetitive sequences, poly T sequence, bar code sequence, invariant portion sequence, promoter sequences, and scaffold sequence more selected from the group consisting further including, method. According to one of Claim 1 to Claim 5, said oligonucleotides are provided which made of 100 to 200 bio-sequence listing, method. According to one of Claim 1 to Claim 5, an oligonucleotide present on RNA target bio-sequence listing Cis - have the same oligonucleotide on guide to guide RNA in action (Cis a-acting), method. According to one of Claim 1 to Claim 5, the method (a) guide RNA encoding a desired target bio-sequence listing including said guide RNA oligonucleotides bio-sequence listing and is introducing a new the [ley which will grow oh library cells including RNA - guide number; (b) said RNA - guide new the [ley which will grow oh number have been introduced DNA obtained from a library cells performing deep using common channel redistribution message; and (c) said guide from the data obtained by deep sequencing RNA - n is and it is burnt frequency including detecting a target sequence, method. According to one of Claim 1 to Claim 5, said RNA - guide number is new the [ley which will grow oh Cpf1 Cas9 protein or protein, method. According to Claim 10, said Cas9 protein is Streptococcus ( According to Claim 10, said Cpf1 protein is candy multi [thwu green onion tax pak ( According to one of Claim 1 to Claim 5, said RNA - guide new the [ley which will grow oh number properties of the, number of PAM sequence RNA - guide new the [ley which will grow oh (i), (ii) RNA - guide new the [ley which will grow oh number target active (on a non-target activity), or (iii) number of RNA - guide new the [ley which will grow oh (off-a target activity) including at least one selected from the group consisting of active target, method. According to Claim 1, said sequencing is to performed deep sequencing (deep sequencing), method. Guide RNA encoding said RNA oligonucleotides including cells including a desired target bio-sequence listing bio-sequence listing and including at least one guide is 2, cell library. Said guide including a guide RNA encoding the desired target RNA oligonucleotides is separated bio-sequence listing and bio-sequence listing including vector. According to Claim 16, said vector is a viral vector, vector. According to Claim 16, said vector lentiviral vector, which is one selected from the group consisting of retroviral vector and plasmid vector, vector. Guide RNA encoding a desired target bio-sequence listing including said guide RNA oligonucleotides is separated bio-sequence listing and including at least one library of vectors including 2 vector. Guide RNA coding bio-sequence listing including a separate oligonucleotide bio-sequence listing and target. 2 Oligonucleotides according to Claim 20 including at least one oligonucleotide library. (A) new the [ley which will grow oh guide target RNA - [thing it does, character number involves setting a target bio-sequence listing; (b) setting said target bio-sequence listing complementary chain with a base forming a gear pair, guide RNA nucleotide sequence encoding design step; (c) said guide RNA oligonucleotide including a designing step bio-sequence listing the purpose and target; said step (a) to (c) and (d) repeating step 1 or more times including including, according to Claim 20 method for constructing a library of oligonucleotides. According to Claim 22, (c) or said step (d) further comprises the step designed synthetic oligonucleotides including, method. In PAM (proto-a spacer provided adjacent Motif) sequence CTTA TTTV or adjacent to, target bio-sequence listing complementary chain (complementary strand) with a base pair sequence including form, separate guide RNA. According to Claim 24, said separated-out number is new the [ley which will grow oh Cpf1 protein RNA - guide for use with guide RNA, RNA isolated guide. According to Claim 25 number 24 anti or separated guide RNA, or nucleic acids encoding the same including, for correcting dielectric compositions. According to Claim 25 number 24 anti or separated guide RNA, or nucleic acid encoding the same; and including nucleic acids encoding the same protein or Cpf1, system for calibrating a dielectric in mammalian cells. According to Claim 24 guide RNA or nucleic acid encoding the same; and Cpf1 protein or nucleic acid encoding the same sequentially or simultaneously introducing isolated mammalian cell including, dielectric correction in an optical method Cpf1 in mammalian cells. Primer Sequence (5 '-3') Lenti_gRNA_Puro cloning FP1 CAC CGG AGA CGT TGA CTA TCG TCT CGC TAC TCT ACC ACT TGT ACT TCA GCG GTC A (sequence number 6) RP1 AAG CTG ACC GCT GAA GTA CAA GTG GTA GAG TAG CGA GAC GAT AGT CAA CGT CTC C (sequence number 7) FP2 GCT TAC TCG ACT TAA CGT GCA CGT GAC ACG TTC TAG ACC GTA CAT GCT TAC ATG GGA TGA (seq ID no 8) RP2 AGC TTC ATC CCA TGT AAG CAT GTA CGG TCT AGA ACG TGT CAC GTG CAC GTT AAG TCG AGT (seq ID no 9) AsCpf 1 oligo library amplification FP ATT TCT TGG CTT TAT ATA TCT TGT GGA AAG GAC GAA ACA CCG TAA TTT CTA CTC TTG TAG (sequence number 10) LbCpf 1 oligo library amplification FP TTT CTT GGC TTT ATA TAT CTT GTG GAA AGG ACG AAA CAC CGT AAT TTC TAC TAA GTG TAG (sequence number 11) As/LbCpf 1 oligo library amplification RP GAG TAA GCT GAC CGC TGA AGT ACA AGT GGT AGA GTA GAG ATC TAG TTA CGC CAA GCT (seq ID no 12) Targeted deep sequencing FP ACA CTC TTT CCC TAC ACG ACG CTC TTC CGA TCT CTT GTG GAA AGG ACG AAA CAC C (sequence number 13) RP GTG ACT GGA GTT CAG ACG TGT GCT CTT CCG ATC TTT GTG GAT GAA TAC TGC CAT TTG TC (sequence number 14) Index of Illumina (indexing) FP (Sequence number 15) AAT GAT ACG GCG ACC ACC GAG ATC TAC AC - (8bp barcode sequence) (sequence number 16) - ACA CTC TTT CCC TAC ACG AC RP (Sequence number 17) CAA GCA GAA GAC GGC ATA CGA GAT - (8bp barcode) - GTG ACT GGA GTT CAG ACG TGT (seq ID no 18) QPCR for WPRE FP GAT ACG CTG CTT TAA TGC CTT TG (seq ID 19) RP GAG ACA GCA ACC AGG ATT TAT ACA AG (sequence number 20) QPCR for FP GCT GTC ATC TCT TGT GGG CTG T (sequence number 21) RP ACT CAT GGG AGC TGC TGG TTC (seq ID no 22) Endogenous target 1 - 5 FP TTG CTG TGG CAG AGC CAG CG (seq ID no 29) RP TTG CTT CAC TTT AAT CCT TTC TTG CAG (seq ID no 30) Endogenous target 6 - 10 FP CTC CTG CAA GAA AGG ATT AAA GTG (seq ID no 31) RP ACC TAC CTA ATA GTT ACT TCC TGA AGG G (sequence number 32) Endogenous target 11 - 14 FP CTC GTT CTT TCC ATC AAA TAG TGT GGT G (seq ID no 33) RP CTG CAG TAA TTG TTA CTC TGT GTC TTC C (seq ID no 34) Endogenous target 15 - 17 FP TTG AGC TGA CCC ATA AAT ACA ACA GG (sequence number 35) RP CCC TCT TAA CTG GAT CAG CAA CGG (seq ID no 36) Endogenous target 18 FP TGG GGT CGC CAT TGT AGT TCC C (sequence number 37) RP GTC ACA AAG ATC AGC ATC AGG CAT GG (seq ID no 38) Endogenous target 19 - 22 FP CGT TCA CCT GGG AGG GGA AG (sequence number 39) RP TCT GCA AAG AAC TTT ATT CCG AGT AAG C (seq ID no 40) Endogenous target 23 - 28 FP CCC AAA AGA CAT ATT CAC CCA GAA TCC C (sequence number 41) RP CAA CAT CAA GGT GTG GGC AGG GCT GC (seq ID no 42) Endogenous target 29 - 30 FP ACC TGG AGT CTG CAG AGC TGG (seq ID no 43) RP AAG CGG TAA ACA AAG GAT AGC TGG (seq ID no 44) Endogenous target 31 - 35 FP CCA TGG GAA ACG AAT ACA GGT CTC G (seq ID no 45) RP CTT CAG AAG AAA AAC CTC CAC TC (sequence number 46) Endogenous target 36 - 37 FP AAC TGA GAA ACA GCC AGA GAG GAA G (seq ID no 47) RP CAT CTG ATG CTG ACT CAG AGC GC (seq ID no 48) Endogenous target 38 - 42 FP GCT GCC ACC CCC TGC TC (seq ID no 49) RP ATC AGA ATG AAA AAT CTC ACC CCT CC (seq ID no 50) Endogenous target 43 - 46 FP GTC TCC GTG ATG GGG GTG G (sequence number 51) RP CTG CCT TGT AAG ACT TTA AAT ATT CTG CTC C (seq ID no 52) Endogenous target 47 - 48 FP AAG CCA TAT TCA GTT TTA GGG AAA AGC (seq ID no 53) RP ATT TCC AAG TAA GCT GCA AGG AAA GC (seq ID no 54) Endogenous target 49 - 52 FP AAG TCT TAC AAG GCA GAG TAA AGA TC (seq ID no 55) RP GCA GGG TAA AAC AAT CGG ACC (seq ID no 56) Endogenous target 53 - 57 FP CAA CCA CCT CAG AAG AGC CAG ATT CC (seq ID no 57) RP CTC TGT AGT TAT TTG AGC AAT GCC AC (seq ID no 58) Endogenous target 58 - 64 FP CAG TGA ATA TAC AGG ATT GGG GTT GTG (seq ID no 59) RP ACA ACT GGT AAG GTG GGC CCA GG (seq ID no 60) Endogenous target 65 - 72 FP CAA GCA CAA ACA AAT CAG GCT AAA TCC (seq ID no 61) RP CCC TGA GCT TGG GGG AGA GTT AC (seq ID no 62) Endogenous target 73 - 78 FP TCC TCT GGG GAA AGA GTG GCC (seq ID no 63) RP TGT GGG GTC GTT CCT GAT GAA AC (seq ID no 64) Endogenous target 79 - 82 FP AAC TGG TTT AGC TAG TGC ATA CAT GC (seq ID no 65) RP GGT GGG AGT TTC TGT TAC AGG CAA C (seq ID no 66) Object Pair (pair) be the minimum read per In assays allow a maximum frequency number is and it is burnt and a sense background AsCpf 1 PAM confirmation 100 8% LbCpf 1 PAM confirmation 30 8% AsCpf 1 target effect profiling 100 8% AsCpf 1 non-target effect profiling 100 8% Is and it is burnt time dependent frequency analysis 300 8% Guide RNA segment of the non-target effect profiling 300 8% Primer Sequence (5 '-3') SpCas 9 oligo library amplification FP TTG AAA GTA TTT CGA TTT CTT GGC TTT ATA TAT CTT GTG GAA AGG ACG AAA CAC C (seq ID no 25) RP TTT CAA GTT GAT AAC GGA CTA GCC TTA TTT TAA CTT GCT ATT TCT AGC TCT AAA AC (seq ID no 26) Targeted deep sequencing (SpCas 9) FP ACA CTC TTT CCC TAC ACG ACG CTC TTC CGA TCT TGG ACT ATC ATA TGC TTA CCG TAA CTT G (seq ID no 27) RP GTG ACT GGA GTT CAG ACG TGT GCT CTT CCG ATC TTT TGT CTC AAG ATC TAG TTA CGC CAA G (seq ID no 28) Category (object) The number of designed pair The filtering conditions The number of filtered pair PAM sequences used Number of other guide sequence design The number of filtering before the other guide sequences AsCpf 1 determining of PAM 1,540 100 Or more read (read), 8% hereinafter is and it is burnt (Background indels) background 1,074 70 Different types 22 18 LbCpf 1 determining of PAM 1,540 30 Or more read (read), 8% hereinafter is and it is burnt (Background indels) background 940 70 Different types 22 16 AsCpf 1 active 2,381 100 Or more read (read), 8% hereinafter is and it is burnt (Background indels) background 1,251 ATTTA 2,381 1,251 The activity of RNA using AsCpf 1 cut (truncated) guide 420 300 Or more read (read), 8% hereinafter is and it is burnt (Background indels) background 315 ATTTA 7 7 Aminopeptidase activity of target out AsCpf 1 2,580 100 Or more read (read), 8% hereinafter is and it is burnt (Background indels) background 1,543 ATTTA 4 3 Aminopeptidase activity of target out LbCpf 1 1,342 30 Or more read (read), 8% hereinafter is and it is burnt (Background indels) background 742 ATTTA 4 Insufficient number analysis does not read In a comparison of biological simulation is and it is burnt frequency 8,327 100 Or more read (read), 8% hereinafter is and it is burnt (Background indels) background 156 70 Different types 3,794 47 Cpf1 delivery method is and it is burnt frequency comparison of different liver 8,327 200 Or more read (read), 8% hereinafter is and it is burnt (Background indels) background 233 70 Different types 3,794 49 Category AsCpf 1 LbCpf 1 Oligonucleotide population new the [ley which will grow mote [tu Cell library Oligonucleotide population new the [ley which will grow mote [tu Cell library The number of designed pair 8,327 3,634 The number of included pair (1 or more read) 8,313 8,146 3,626 3,497 Percentage (%) n/designed included 99. 8% 97. 8% 99. 8% 96. 2% The number of deep [...] contributed by reading 1,238,978 10,378,634 475,610 584,771 Category Traditional die Of the present inventionmethod Procedure Cost (USD) Labor (unit) Procedure Cost (USD) Labor (unit) Oligonucleotide synthesis Synthesis 54,000 - Synthesis 2,200 - Library number bath Phosphorylation 4,480 100 Oligonucleotide library amplification 53 0. 5 Junction (Ligation) 128 20 [...] assembly 159 0. 5 Transformation and plating - 220 Transformation and plating 165 1 Plasmid number bath and sequencing 30,000 200 Plasmid number bath and cell library number bath 112 3 CRISPR provided Cpf1 delivery Transfected 749 100 Transduction - 1 Deep sequencing preparing samples Genome DNA separation - 500 Genome DNA separation - 2 PCR for deep common channel redistribution message - 100 PCR for deep common channel redistribution message - 1 Chlorine 89,357 1,240 2,689 9