Method for screening of a lipase having improved enzymatic activity using yeast surface display vector and the lipase
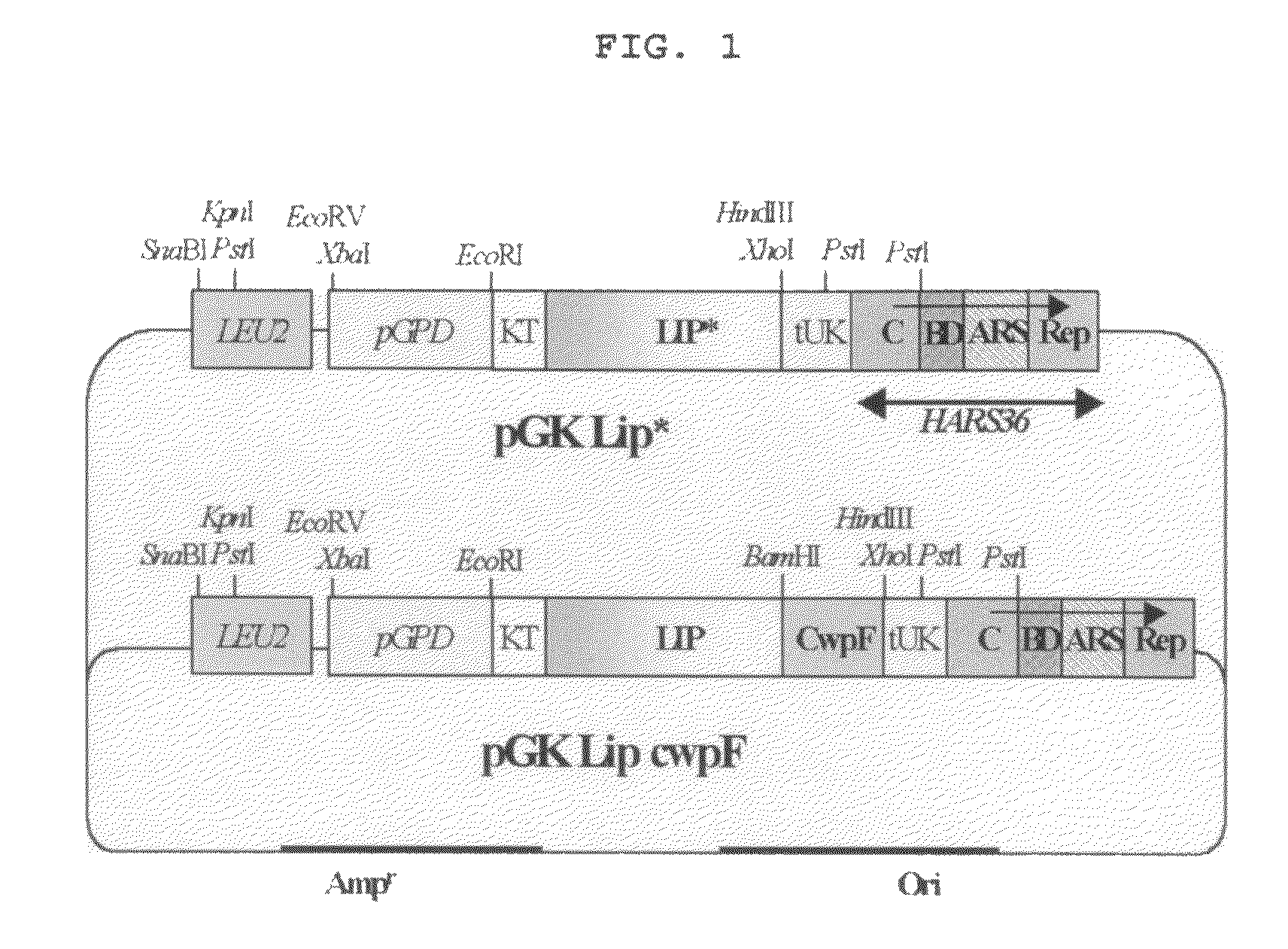
The present invention relates to a method for screening of a lipase having an improved enzymatic activity using a yeast surface display vector and the mutant lipase prepared by the same, more particularly to the method comprising; 1) cloning lipase gene into a surface display vector, 2) preparing a mutant gene library of lipase of the step 1 by mutagenic PCR, 3) transforming the mutant lipase gene library of the step 2 and a surface display vector into a host cell, and 4) measuring the activity of the mutant lipase displayed in the surface of the transformed host cell and select the mutant lipase with evolved activity, and the mutant lipase prepared by the same. Lipase is a carboxylic ester hydrolase and is widely used for food and surfactant industry as well as the synthesis of various chiral compounds. Among many microorganism-originated lipases, CALB is industrially very important in the selection of optical isomers and in the synthesis of polyester (Anderson et al., Recently, studies to express foreign proteins on the cell surface of unicellular organism such as yeast, bacteria including bacteriophage, etc. have been actively undergoing and applied for the production of a new vaccine, the screening of antigens and antibodies, the fixation of useful enzymes onto cell surfaces, and so on. For example, researches on the expression of proteins on cell surface have been progressed using a kind of yeast ( In addition, according to a recent report, the development of industrially effective biocatalyst comes true by the simultaneous expression of various enzymes in a cell (Murai et al., Recently, studies on the screening method using The present inventors isolated novel cell wall attachment-mediating proteins from the industrial yeast, In order to mass-produce lipase, a useful biological catalyst, the present inventors have constructed The present invention provides a method for screening of a mutant lipase having an improved enzymatic activity comprising the steps of 1) cloning a lipase gene into a surface display vector, 2) preparing a mutant lipase gene library by mutagenic PCR using the lipase gene in the expression vector of the step 1 as a template, 3) transforming the mutant lipase gene library of the step 2 and the surface display vector into host cells, and 4) measuring the activity of the mutant lipase displayed in the surface of the transformed host cell and selecting the lipase with improved activity from the mutant pool. The present invention also provides a mutant lipase protein prepared by the screening method of the present invention, in which #219 leucine and/or #278 leucine of The present invention also provides a gene coding the mutant lipase protein. The present invention also provides an expression vector containing the above gene. The present invention also provides a transformant in which the above expression vector was introduced. The present invention also provides a method for the production of the mutant lipase protein by cultivation of the above transformant. Further features of the present invention will appear hereinafter. The present invention relates to a method for screening of a mutant lipase having an improved enzymatic activity comprising the following steps: 1) Cloning a lipase gene into a surface display vector; 2) Preparing a mutant lipase gene library by mutagenic PCR using the lipase gene in the surface display vector of the step 1 as a template; 3) Transforming the mutant lipase gene of the step 2 and the surface display vector into host cells; and 4) Measuring the activity of the mutant lipase displayed in the surface of the transformed host cell selecting the lipase with improved activity from the mutant pool. The ‘surface display vector’ in this invention means a vector expressing a foreign protein stably on a cell surface. In this invention, The surface display vector used for the screening in this invention is the vector expressing a foreign protein on the surface of a transformant and is characterized by including a promoter gene, a gene coding a secretion signal sequence, a lipase gene or a mutant lipase gene, a surface display mediating gene and a terminator gene. It is preferable to select a promoter gene from a group consisting of GAPDH, PGK, ADH, PHO5, GAL1, GAL10, SED1, MOX, TEF and TPI, select a gene coding a secretion signal sequence from a group consisting of Mf α, PHO6, SUC2, AMY, SED and killer toxin, and select a surface display mediating gene, a factor that can express lipase on the cell surface, from a group consisting of SED1, PIR2, TIP1, CWP1, GAS1 AND WSC1, but not always limited thereto. As a host cell for the transfection in step 3 of the screening method in this invention, yeasts such as In the preferred embodiments of the present invention, the present inventors used the surface display vector that was introduced into the transformants (Accession Nos: KCTC 0824BP, KCTC 0825BP, KCTC 0826BP, KCTC 0827BP and KCTC 0828BP) that were deposited at Gene Bank of Korea Research Institute of Bioscience and Biotechnology (Korea Patent Application No: 2000-42939). According to the screening method of the present invention, PCR was performed to induce mutation of a lipase gene after cloning the gene into the surface display vector. Host cells were transfected with the mutant gene. Then, the mutant gene was expressed in host cells. As a result, the mutant lipase was located on the surface of the transformant. Thus, the mutant lipase having high activity can be screened readily and fast by measuring the activity of the mutant lipase expressed on the surface of the transformant. The present invention also provides a mutant lipase protein prepared by the screening method of the present invention, in which #219 leucine and/or #278 leucine of The present invention provides a mutant protein in which #219 amino acid and/or #278 amino acid of The hydrophobic amino acid leucine of the mutant protein of the present invention that is #219 amino acid of CALB represented by SEQ. ID. No 14 is preferably replaced by a hydrophilic amino acid selected from a group consisting of glutamine, histidine, arginine, lysine, serine, threonine, asparagine and glutamic acid, and is more preferably substituted by glutamine, leading to be represented by SEQ. ID. No 11. Also, #278 amino acid leucine of CALB represented by SEQ. ID. No 14 is preferably replaced by an amino acid selected from a group consisting of proline, tyrosine, phenylalanine, tryptophane and valine, and is more preferably substituted by a proline, making it be represented by SEQ. ID. No 9. Also, #219 amino acid leucine of CALB represented by SEQ. ID. No 14 is preferably replaced by a hydrophilic amino acid selected from a group consisting of glutamine, histidine, arginine, lysine, serine, threonine, aspartic acid and glutamic acid and at the same time, #278 amino acid, leucine, is preferably replaced by an amino acid selected from a group consisting of proline, tyrosine, phenylalanine, tryptophane and valine. And #219 amino acid and #278 amino acid are more preferably substituted by glutamine or proline, respectively, in the same protein, making it be represented by SEQ. ID. No 10. #219 amino acid leucine of CALB is a hydrophobic amino acid exposed on the surface. When the above leucine is replaced by a hydrophilic amino acid, the stability in water increases, leading to the increase of the enzyme activity of a mutant protein. #278 amino acid leucine of CALB is located at 10thα-helix of CALB that has been reported to work as a lid for lipase active site (Uppenberg et al., The enzyme activity was measured with cell culture supernatants of a transformant producing a secreted form of mutant lipase protein. As a result, the affinities to substrates of the mutant proteins having amino acid sequences represented by SEQ. ID. No 9, No 10 and No 11 were similar each other, but had 5 times, 10 times and three times higher enzyme activity respectively than a wild type CALB (see Table 3). The enzyme activity was also measured with whole cell fraction of the transformant expressing the mutant lipase protein of the present invention on the surface. As a result, the mutant proteins having amino acid sequences represented by SEQ. ID. No 9 and No 10 showed 5 times higher activity than a wild type CALB (see Table 1). Thus, the mutant lipase protein of the present invention is confirmed to have higher enzyme activity than a wild type lipase protein. The present invention also provides a gene coding the mutant lipase protein. It is preferable that the gene of the present invention is coding a mutant protein in which #219 amino acid of CALB is replaced by a hydrophilic amino acid selected from a group consisting of glutamine, histidine, arginine, lysine, serine, threonine, aspartic acid and glutamic acid, and is more preferable that the gene is represented by SEQ. ID. No 8. It is also preferable that the gene of the present invention is coding a mutant protein in which #278 amino acid of CALB is replaced by an amino acid selected from a group consisting of proline, tyrosine, phenylalanine, tryptophane and valine, and is more preferable that the gene is represented by SEQ. ID. No 6. It is also preferable that the gene of the present invention is coding a mutant protein in which #219 amino acid of CALB is replaced by a hydrophilic amino acid selected from a group consisting of glutamine, histidine, arginine, lysine, serine, threonine, aspartic acid and glutamic acid, and #278 amino acid is substituted with an amino acid selected from a group consisting of proline, tyrosine, phenylalanine, tryptophane and valine. And it is more preferable that the gene is represented by SEQ. ID. No 7. The present invention also provides an expression vector including the said gene. The expression vector of the present invention includes a gene coding the mutant lipase protein of the invention, and is prepared to express the mutant lipase protein on the cell surface. The vector expressing the mutant lipase protein of the present invention is characterized by having a promoter gene, a gene coding a secretion signal sequence, a mutant lipase gene, a surface display mediating gene and a terminator gene. The promoter gene of the surface display vector is preferably selected from a group consisting of GAPDH, PGK, ADH, PHO5, GAL1, GAL10, SED1, MOX, TEF and TPI, and the gene coding a secretion signal sequence is preferably selected from a group consisting of MF-α, PHO5, SUC2, AMY, SED and killer toxin. The surface display-mediating gene is a factor to express lipase on the cell surface, and is preferably selected from a cell wall constituting gene group consisting of SED1, PIR2, TIP1, CWP1, GAS1, and WSC1. However, the choice is not always limited thereto. In the preferred embodiments of the present invention, the present inventors constructed an expression vector having base sequences represented by SEQ. ID. No 6, No 7 or No 8 which are the genes coding the mutant protein of the present invention. The present invention also provides a transformant prepared by introducing the said surface display vector into host cells. Particularly, the transformant of the present invention was prepared by introducing the said expression vector including the mutant lipase gene into host cells. Host cells for the present invention can be selected from yeasts such as In the preferred embodiments of the present invention, yeast was used as a host cell. Particularly, The present invention also provides a method for producing a mutant lipase protein by culturing the said transformant. The transformant of the present invention is preferably cultured at 2° C.-20° C. lower temperature than usual host cell culturing temperature to produce the protein. Particularly, In addition, after mass-culturing the transformant of the present invention by fed-batch culture, it was confirmed that cell growth and the activity of lipase in cell culture solution were excellent (see The application of the preferred embodiments of the present invention is best understood with reference to the accompanying drawings, wherein: pGPD: GAPDH promoter, KT: Killer toxin secretion signal sequence, tUK: Unknown terminator, c: Terminal region of HARS36, BD: Bent DNA domain of HARS36, ARS: Self-replicable sequence of HARS36, Rep: Telomeric repeated sequence of HARS36. A: Active site structure of wild type lipase, B: Active site structure of mutant lipase. Lane 1: Wt; wild type Lane 2: Lip-CwpF; a strain containing a vector displaying lipase on the surface (pLGK-Lip-CwpF), Lane 3 and Lane 4: Lip10-CwpF and Lip14-CwpF; strains containing the surface display vector of Lip10 and Lip14, Lane 5: Lip*; a strain containing a secretion vector of lipase (pLGK-Lip*), Lane 6 and Lane 7: Lip10 and Lip14; strains containing the secretion vector of mutant lipase Lip10 and Lip14 (pLGK-Lip*). Lane 1: Lipwt* strain, Lane 2: Lip10* strain, Lane 3: Lip14* strain, Lane 4 and Lane 5: LP mutant strain, Lane 6 and Lane 7: LQ mutant strain, Lane 8 and Lane 9: LPQ mutant strain. Practical and presently preferred embodiments of the present invention are illustrative as shown in the following Examples. However, it will be appreciated that those skilled in the art, on consideration of this disclosure, may make modifications and improvements within the spirit and scope of the present invention. The present inventors constructed a vector expressing CALB on the surface of Particularly, in order to construct a vector to secrete CALB to the media, CALB gene was first obtained from In order to prepare a vector expressing CALB on the cell surface, CALB gene was first obtained from As a result, the expression of CALB was confirmed by the presence of the active ring around the transformant. In order to establish CALB mutant library, the present inventors used in vivo recombination, which was once reported in In vivo recombination is the method in which cells are transfected with a vector fragment and a synthesized insert fragment having homology with the vector in both ends of the DNA fragment together, so that an authentic circular vector is produced by the recombination therein. The in vivo recombination is simple and efficient method with no need to establish a library in advance in The pGK-Lip-CwpF, a vector expressing CALB on the cell surface, was digested with EcoRI/PstI, resulting in the obtainment of 5 kb fragment. The fragment was recovered through gel and used as a vector fragment for the transformation. PCR was performed using PCR premix kit (Bioneer, Korea) with primers represented by SEQ. ID. No 4 and No 5 prepared by the cross of the vector fragment and 100 bp of HARS 36 region at 94° C. for 3 minutes and 25 cycles of denaturation at 94° C. for 30 seconds, annealing at 55° C. for 30 seconds, and extension at 72° C. for 1 minute, followed by a final extension at 72° C. for 7 minutes. At this time, pGK-Lip-CwpF was used as a substrate. At last, 100 ng of the resultant fragment was used for the transformation of The transformed strains were randomly selected to measure the lipase activity on YPD flat medium containing 1% tributyline, leading to the confirmation of constructed library having a uniform activity. In order to construct CALB mutant Library, the present inventors used error-prone PCR and in vivo recombination. Particularly, error-prone PCR was performed with primers represented by SEQ. ID. No 4 and No 5 using pGK-Lip-CwpF, a vector expressing CALB on the cell surface, as a template. PCR random error-prone kit (Clontec, USA) was used to induce 2-5 errors per 1 kb. DNA fragments were recovered after being cut by required size on gel. Amplification PCR was performed with those fragments by using PCR premix kit (Bioneer, Korea) with primers represented by SEQ. ID. No 4 and No 5 at 94° C. for 3 minutes and 25 cycles of denaturation at 94° C. for 30 seconds, annealing at 55° C. for 30 seconds, and extension at 72° C. for 1 minute, followed by a final extension at 72° C. for 7 minutes. 100 ng of the fragment obtained by the above process and 100 ng of the 5 kb vector fragment obtained by digesting pGK-Lip-CwpF with EcoRI/PstI were mixed and transfected into As a result, about 1×104transformed strains were obtained. The present inventors selected CALB mutant having high lipase activity from the CALB mutant library constructed in the above <Example 3>. About 7,000 strains of the mutant library were inoculated on YPD plate medium containing 1% tributyline and cultured for 24 hours. Then, 23 strains showing large activity circle were primarily screened. The primarily selected strains were inoculated in YPD liquid medium and cultured at 37° C. for 16 hours, followed by centrifugation at 5,000 rpm for 5 minutes to separate cell fraction and culture supernatant. The cell fraction was suspended in 50 mM tris buffer solution (pH 7.5), which was washed and then suspended again with the same buffer solution by the same amount. In order to measure the lipase activity, ρ-nitrophenyl palmitate (referred as ‘pNPP’ hereinafter) was used as a substrate. Reaction solution was prepared by mixing 10 μl of 10 mM pNPP, 40 μl of ethyl alcohol and 950 μl of 50 mM tris buffer (pH 7.5), to which 100 μl of cell suspension was added. After the reaction for 2 hours at 25° C., OD450was measured. The method to measure the lipase activity using pNPP as a substrate is to measure the extinction level of pNP group at 405 nm. The pNP group is isolated from pNPP (pNP group and palmitate were combined therein) by lipase. In this invention, 1 unit of lipase activity is defined to be the enzyme activity isolating 1 uM of pNP group per 1 minute. As a result, strains Lip14, Lip10 and Lip23, which all had excellent activity in whole cell fraction, were finally selected. The present inventors analyzed the mutant genes of the three mutant strains obtained in the above <Example 4>. In order to recover the CALB mutant gene, the genome of the mutant strain was first purified, which was used as a template. PCR was performed using PCR premix kit (Bioneer, Korea) with primers represented by SEQ. ID. No 4 and No 5 at 94° C. for 3 minutes and 25 cycles of denaturation at 94° C. for 30 seconds, annealing at 55° C. for 30 seconds, and extension at 72° C. for 1 minute, followed by a final extension at 72° C. for 7 minutes. DNA fragments were recovered from gel and the DNA sequences were analyzed. As a result, DNA sequences of Lip10 and Lip14 were each represented by SEQ. ID. No 6 and No 7, and their corresponding amino acid sequences were represented by SEQ. ID. No 9 and No 10 respectively. And it was confirmed that the 278thleucine of Lip10 and Lip14 was replaced by proline, and additionally the 219thleucine of Lip14 was substituted with glutamine ( The 278thleucine is located at the 10thα-helix of CALB and the region has been reported as a lid structure of the lipase active site (Uppenberg et al., The present inventors measured the enzyme activity of lipase using CALB mutant strain selected in the above <Example 4>. In order to construct a vector to express a gene on the cell surface, genomes of mutant strain and wild type CALB-expressing strain were isolated, which were used as templates for PCR. PCR was performed using PCR premix kit (Bioneer, Korea) with primers represented by SEQ. ID. No 4 and No 5 at 94° C. for 3 minutes and 25 cycles of denaturation at 94° C. for 30 seconds, annealing at 55° C. for 30 seconds, and extension at 72° C. for 1 minute, followed by a final extension at 72° C. for 7 minutes. The obtained DNA fragments were digested with EcoRI/ClaI and were inserted into a single copy integration vector ‘AMIpLD1’ (Agaphonov et al., In order to construct a vector to secrete a gene product to the media, genomes of mutant strain and wild type strain were isolated, which were used as templates for PCR. PCR was performed using PCR premix kit (Bioneer, Korea) with primers represented by SEQ. ID. No 1 and No 2 at 94° C. for 3 minutes and 25 cycles of denaturation at 94° C. for 30 seconds, annealing at 55° C. for 30 seconds, and extension at 72° C. for 1 minute, followed by a final extension at 72° C. for 7 minutes. The obtained DNA fragments were inserted into EcoRI/BamHI site of AMIpLD1 along with GAPDH promoter to construct vectors pLGK-Lip10* and pLGK-Lip14*. After confirming the introduction of wild type and mutant CALB in the vector by the DNA sequencing, As a result, 5 times increased lipase activity was observed in two kinds of mutant strains when they displayed on the cell surface. When the lipase was secreted out of cells, the activity in culture supernatant was confirmed to be increased 5 times (Lip10) and 10 times (Lip14) respectively, comparing to the wild type (Table 1). Also, the culture supernatant of strain secreting CALB was investigated by 12% SDS-PAGE and the results were confirmed by silver staining ( The present inventors separated and purified wild type CALB and mutant CALB for the accurate analysis of the characteristics of the mutant CALB. Each strain secreting CALB was cultured in YPD medium for 18 hours, followed by centrifugation at 5,000 rpm for 5 minutes to obtain culture supernatant. The culture supernatant was 10-fold concentrated by ultrafiltration. After then, ammonium sulfate was added into the protein solution until the concentration of ammonium sulfate was adjusted to 1 M. Let the recovered solution pass through butyl sepharose CL-4B column (Pharmacia, USA) saturated with 50 mM, pH 6.5 phosphate buffer containing 1 M ammonium sulfate. The concentration was lowered by 100% water to elute proteins. Among the eluted protein fractions, only the fractions showing lipase activity were recovered, which were concentrated again by ultrafiltration, and then, purified by using superdex-G200 gel filtration column chromatography (Pharmacia, USA). The purified Lip10, Lip14 and wild type lipase (Lip wt) were-obtained. The mutant protein and the wild type protein were compared by measuring Km and Kcat with the purified proteins when pNPP was used as a substrate. 10 μl of protein solution was added to lipase reaction solution prepared by supplementing pNPP at serial different concentration. The optical density of the isolated pNP group was measured at 405 nm to determine Vi. Km value of each protein was calculated by applying Michales-Menten equation using the Vi. As a result, Km values of both Lip10 and Lip14 were not increased. On the other hand, Kcat value of each protein was increased more than 6.5 times, suggesting that the activity of Lip10 and Lip14 was increased without changing the affinity to substrate (Table 2). The remaining activity of the protein was also measured after letting the protein at 50° C. for 10 minutes to test the heat stability. As a result, there was no big difference between the mutant protein and the wild type protein in heat stability. Stereoselectivity was also measured by using (R,S)-acetyl ester as a substrate. As a result, there was no big difference between the mutant protein and the wild type protein in stereoselectivity. So, each mutant protein was confirmed to have 6-fold increased enzyme activity without damaging the stability and specificity of the protein. The present inventors investigated the mutation characteristics by using site-directed mutagenesis to confirm whether the characteristic change of mutant protein was solely caused by the change of amino acid. L278P (the 278thleucine was replaced by proline, which was present both in Lip10 and Lip14) mutation and L219Q (the 219thleucine was replaced by glutamine, which was present only in Lip14) mutation were induced by PCR for the site-directed mutagenesis in wild type gene, and analyzed their effect. For the site-directed mutagenesis, PCR was performed using pfu polymerase and wild type CALB gene as a substrate with primers represented by SEQ. ID. No 15, No 16, No 17 and No 18 at 94° C. for 3 minutes and 15 cycles of denaturation at 94° C. for 1 minute, annealing at 55° C. for 1 minute, and extension at 72° C. for 1 minute, followed by a final extension at 72° C. for 10 minutes. In order to synthesize L278P mutant gene, one gene was prepared by PCR with primers represented by SEQ. ID. No 2 and No 17 using wild type gene as a substrate. The other gene was also synthesized by PCR with primers represented by SEQ. ID. No 4 and No 18. The two synthesized DNA fragments were mixed, and the mixture was used as a substrate for PCR with primers represented by SEQ. ID. No 2 and No 4, resulting in the connection of the two gene fragments. At that time, 5 μl of each gene fragment and 3 μl of each primer were added into the PCR reaction mixture, and then PCR was performed using ExTaq polymerase (Dakara, Japan) at 94° C. for 3 minutes and 20 cycles of denaturation at 94° C. for 30 seconds, annealing at 55° C. for 30 seconds, and extension at 72° C. for 90 seconds, followed by a final extension at 72° C. for 7 minutes. The gene synthesized by the above method was digested with EcoRI/ClaI, which was inserted into the corresponding region of the vector ‘pLGK-Lip-CwpF’ and then named ‘pLGK-LP’. In order to synthesize L219Q mutant gene represented by SEQ. ID. No 8, one gene was prepared by PCR with primers represented by SEQ. ID. No 2 and No 15 using wild type gene as a substrate. The other gene was also synthesized by PCR with primers represented by SEQ. ID. No 4 and No 16. The two synthesized DNA fragments were mixed, and the mixture was used as a substrate for PCR with primers represented by SEQ. ID. No 2 and No 4, resulting in the connection of the two gene fragments. At that time, 5 μl of each gene fragment and 3 μl of each primer were added-into the PCR reaction mixture, and then PCR was performed using ExTaq polymerase (Dakara, Japan) at 94° C. for 3 minutes and 20 cycles of denaturation at 94° C. for 30 seconds, annealing at 55° C. for 30 seconds, and extension at 72° C. for 90 seconds, followed by a final extension at 72° C. for 7 minutes. The gene synthesized by the above method was digested with EcoRI/ClaI, which was inserted into the corresponding region of the vector ‘pLGK-Lip-CwpF’ and then named ‘pLGK-LQ’. In order to obtain a gene inducing both L278P mutation and L219Q mutation, PCR was performed with primers represented by SEQ. ID. No 2, No 15, No 4 and No 16 using pLGK-LP gene as a substrate, resulting in the construction of ‘pLGK-LPQ’ following the same process as the above. As a result, LP and LPQ showed similar increased activity as Lip10 and Lip14 (Table 3). LQ that was composed of amino acids represented by SEQ. ID. No 11 showed three times higher lipase activity than the wild type gene. Culture supernatant of each strain was compared by SDS-PAGE to investigate the cause of the increased LQ activity. As a result, LQ produced as many proteins as LPQ ( Thus, it was confirmed that L219Q mutation increased the protein productivity, while L278P mutation decreased the protein activity. Considering Kcat value, Lip14, rather than Lip10, was believed to have the effect of increasing the protein production and it's activity as well, suggesting that L219Q mutation was involved in variety of fields. The present inventors measured CALB activity at various culture temperatures in order to determine the optimum culture condition for CALB. The activity of As a result, the culture supernatant from the cells grown at 25° C. showed the highest activity of lipase and growth. In the meantime, cultivation at 37° C. rather caused rapid decrease of the activity by the lapse of time ( That seemed to be the effect of protease secreted in cells during apoptosis. The experiment was repeated with As a result, it was confirmed that For the optimum production of enzyme, the mutant CALB was expressed in Lip14 gene was synthesized by PCR using primers represented by SEQ. ID. No 2 and No 12, which was then linked to EcoRI/BamHI site of the vector ‘YEGα HIR525’ (Choi et al., As a result, the activity circle was observed on the plate. The confirmed strain was inoculated in YPDG liquid medium and induced to be expressed at 30° C. for 24 hours and the activity was measured. In order to investigate if As a result, when it was expressed at 20° C. (720 AU), the activity was 10 times as high as that when expressed at 30° C. In order to mass-produce improved CALB using As a result, as shown in As explained hereinbefore, the screening method for the mutant lipase of the present invention facilitates the preparation of a transformant producing mutant lipase having an improved enzymatic activity. The mutant lipase prepared from the said transformant can be fixed on cell surface and is reproductive, so that the mass-production is possible. Thus, the method of the present invention that can screen the lipase having an improved enzymatic activity can be effectively used for the various fields, such as food and detergent industry. Those skilled in the art will appreciate that the conceptions and specific embodiments disclosed in the foregoing description may be readily utilized as a basis for modifying or designing other embodiments for carrying out the same purposes of the present invention. Those skilled in the art will also appreciate that such equivalent embodiments do not depart from the spirit and scope of the invention as set forth in the appended claims. The present invention relates to a method for screening of the lipase having improved enzymatic activity using yeast surface display vector and the mutant lipase prepared by the same, more particularly to the method comprising; 1) cloning lipase gene into surface display vector, 2) preparing mutant lipase gene library of the step 1 by mutagenic PCR, 3) transforming the mutant lipase gene library of the step 2 and surface display vector into host cell, and 4) measuring the activity of the mutant lipase displayed in the surface of the transformed host cell and selecting the mutant lipase prepared by the same. The method of the present invention can screen the lipase having improved enzymatic activity. So, it can effectively be used for the various fields, such as food and detergent industry. 1. A mutant lipase protein of 2. The mutant lipase protein as set forth in 3. A polynucleotide encoding the mutant lipase protein of 4. The polynucleotide as set forth in the nucleotide sequence comprises SEQ. ID. No 8. 5. An expression vector comprising the polynucleotide of 6. The expression vector as set forth in 7. A transformant in which the expression vector of 8. A method for producing the mutant lipase protein of 9. A mutant lipase protein of 10. A polynucleotide encoding the mutant lipase protein of 11. An expression vector comprising the polynucleotide of 12. A transformant in which the expression vector of 13. A method for producing the mutant lipase protein of 14. A mutant lipase protein of 15. A polynucleotide, comprising a base sequence comprising SEQ. ID. No 7 coding the mutant lipase protein of 16. An expression vector comprising the polynucleotide of 17. A transformant in which the expression vector of 18. A method for producing the mutant lipase protein of 19. The method as set forth in any of 20. The method as set forth in any of 21. The method as set forth in any of FIELD OF THE INVENTION
BACKGROUND OF THE INVENTION
DETAILED DESCRIPTION OF PREFERRED EMBODIMENTS
BRIEF DESCRIPTION OF THE DRAWINGS
EXAMPLES
Example 1
Preparation of CALB Expressing Vector and Transformant
Example 2
Construction of a Library by In Vivo Recombination
Example 3
Construction of CALB Mutant Library
Example 4
CALB Mutant Selection
Example 5
Analysis of CALB Mutants
Example 6
Measurement of the Activity of CALB Mutant Strains
Activity in culture supernatant(U/l) Lip* 15,255 Lip10* 86,535 Lip14* 164,700 Activity in whole cell fraction (U/l) Lip CwpF 174 Lip10 CwpF 831 Lip14 CwpF 765 Example 7
Purification of CALB from Mutant Strains and Analysis of the Characteristics Thereof
CALB activity of each mutant strain Km(μmol) Vmax Kcat(S-1p mol-1) Lip wt 20.4 1.99 130.9 Lip10 21.4 12.9 850.9 Lip14 23.8 13.7 898.0 Example 8
Analysis of the Mutation Characteristics Using Site-Directed Mutagenesis
Comparison of activity of site-directed mutagenesis- induced CALB Activity in culture Lipase supernatant (U/l) Lip wt 17,458 Lip10 118,401 Lip14 190,854 LP 98,890 LQ 48,850 LPQ 200,300 Example 9
Variation of CALB Expression According to the Culture Temperature
Example 10
CALB Expression in
Example 11
Optimization for the Mass-Production of Improved CALB Using Fermentor
INDUSTRIAL APPLICABILITY






CPC - классификация
CC1C12C12NC12N1C12N15C12N15/C12N15/1C12N15/10C12N15/103C12N15/1037C12N9C12N9/C12N9/1C12N9/18C12QC12Q1C12Q1/C12Q1/6C12Q1/68C4C40C40BC40B4C40B40C40B40/C40B40/0C40B40/02IPC - классификация
AA6A61A61KA61K3A61K38A61K38/A61K38/4A61K38/46CC0C07C07HC07H2C07H21C07H21/C07H21/0C07H21/02C07KC07K1C07K14C07K14/C07K14/0C07K14/00C1C12C12NC12N1C12N1/C12N1/1C12N1/15C12N1/19C12N15C12N15/C12N15/0C12N15/09C12N15/1C12N15/10C12N15/6C12N15/63C12N9C12N9/C12N9/1C12N9/18C12N9/2C12N9/20C12QC12Q1C12Q1/C12Q1/3C12Q1/34C12Q1/6C12Q1/68C12RC12R1C12R1/C12R1/7C12R1/72C12R1/78C4C40C40BC40B4C40B40C40B40/C40B40/0C40B40/02Цитирование НПИ
435/198Kieke, M.C. et al. “Isolation of anti-T cell receptor scFv mutants by yeast surface display”, Protein Engineering, vol. 10(11) pp. 1303-1310, 1997.
Kim et al. A cell surface display system using novel GPI-anchored proteins in Hansenula polymorpha. Yeast 19: 11153-1163. 2002.
Kim et al. A cell surface display system using novel GPI-anchored proteins in Hansenula polymorpha. Yeast 19: 1153-1163. 2002. cover page.
Kim, Y.J. et al. “Effect of Linoleic Acid Concentration on Conjugated Linoleic Acid Production by Butyrivibrio fibrisolvens A38”, Applied and Environmental Microbiology, vol. 66(12) pp. 5226-5230, 2000.
Murai, T. et al. “Development of an arming yeast strain for efficient utilization of starch by co-display of sequential amylolytic enzymes on the cell surface”, Appl Microbiol Biotechnol, vol. 51 pp. 65-70, 1999.
Patkar, S. et al. “Effect of mutations in Candida antarctica B lipase”, Chemistry and Physics of Lipids, vol. 93 pp. 95-101, 1998.
Patkar, S.A. et al. “Effect of mutation in non-consensus sequence Thr-X-Ser-X-Gly of Candida antarctica lipase B on lipase specificity, specific activity and thermostability”, Journal of Molecular Catalysis B: Enzymatic, vol. 3 pp. 51-54, 1997.
Sawano, A. et al. “Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis”, Nucleic Acids Research, vol. 28(16) pp. i-vii, 2000.
Schreuder, M.P. et al. “Targeting of a Heterologous Protein to the Cell Wall of Saccharomyces cerevisiae”, Yeast, vol. 9 pp. 399-409, 1993.
Shiraga, S. et al., “Construction of the combinatorial library of Rhizopus oryzae lipase mutated in the lid domain by displaying on yeast cell surface”, J. Molecular Catalysis B: Enzymatic, vol. 17, issues 3-5, pp. 167-173, (Jun. 7, 2002).
Uppenberg, J. et al. “The sequence, crystal structure determination and refinement of two crystal forms of lipase B from Candida antarctica”, Structure, vol. 2(4) pp. 293-308, 1994.
VanAntwerp, J.J. et al. “Fine Affinity Discrimination by Yeast Surface Display and Flow Cytometry”, Biotech. Prog., vol. 16 pp. 31-37, 2000.